CRISPR’s Tricks: How Cas Enzymes Capture, Trim, and Integrate Foreign DNA
New work from Jennifer Doudna’s lab could be useful for CRISPR-based technologies that seek to repurpose Cas1–Cas2 for molecular recording and information storage
By Jonathan D Grinstein, PhD -June 16, 2023
Credit: luismmolina / iStock / Getty Images Plus
Owen T. Tuck, a graduate student in Jennifer Doudna’s lab at the University of California, Berkeley, thinks that the enzymes used for genome editing and engineering (such as Cas9 and Cas12) receive far too much attention when it comes to CRISPR. In his opinion, they are not the true heroes of the bacterial antiviral system.
The acquired sequence that Cas9 and Cas12 follow in prokaryotic adaptive immunity, the protospacer sequence, determines how the immune system functions. He believes that the CRISPR machinery that does the viral DNA acquisition and insertion is the unsung hero of the CRISPR system.
“CRISPR encodes viral derived short sequences of DNA in a genomic locus called the CRISPR array,” Tuck told GEN. “Those sequences will then guide the interference of the viral genome if it should arise again. It’s the memory of the immune system.”
It is really a question of self versus non-self, he continued. If bacteria acquire a sequence in their CRISPR array that is self-targeting, they will probably kill themselves, and this is a theme across autoimmunity, which should be avoided.
Tuck is interested in how CRISPR-Cas has adapted to identify and incorporate harmful genetic sequences into their genome. CRISPR systems maintain genome integrity and avoid autoimmunity by distinguishing between self and non-self. In some microorganisms, the Cas4 endonuclease assists CRISPR adaptation, but around 40% of CRISPR subtypes lack Cas4.
hose prokaryotes that lack Cas4 often use the Cas1–Cas2 integrase, but it is not well understood how Cas1–Cas2 can tell viral DNA apart from their own DNA.
In an article published in Nature, Tuck and co-first authors Joy Y. Wang and Petr Skopintsev from the Doudna lab, demonstrate that Cas1 comes together with a fusion protein of Cas2, and an exonuclease called DEDDh to form a trimmer-integrase that catalyzes coordinated DNA capture, trimming, and integration of viral sequences into bacterial DNA.
While Tuck’s research interests lie primarily in the basic science of microbial biochemistry, he acknowledges that the team’s work will be useful for CRISPR-based technologies that aim to repurpose Cas1–Cas2/DEDDh for in vivo recording of molecular events, such as capturing the order of transcriptional events.
The two-step CRISPR dance
The specific question that the Doudna lab research team was able to answer is the role of these exonucleases in CRISPR adaptive immunity. “What do these trimmers do?” asked Tuck. “How do they handle this process that needs to be quite precise, involving the trimming of a substrate to a very short length and temporal gating of a certain sequence called the PAM (protospacer adjacent motif)?”
What they uncovered was a delicate dance of two activities: ruler-guided trimming and PAM protection.
The ruler-guided section of the trimming process is used to reduce the size of the foreign DNA until it is tolerable to the host. They discovered that the fusion of Cas1 and Cas2 can determine the size of the protospacer that will be inserted into the CRISPR array and then preserve and protect the length of the foreign sequence. Finally, the Cas1–Cas2/DEDDh fusion attaches a PAM that functions as an immune system tag.
However, this tag must be removed before the CRISPR array can be inserted. Otherwise, the bacteria will mistakenly target its own DNA. The scientists in the Doudna lab used cryogenic electron microscopy (cryo-EM) to show that the PAM sequence is released by Cas1 and cleaved by the Cas2/DEDDh exonuclease prior to genome integration, marking inserted DNA as self and preventing aberrant CRISPR targeting of the host.
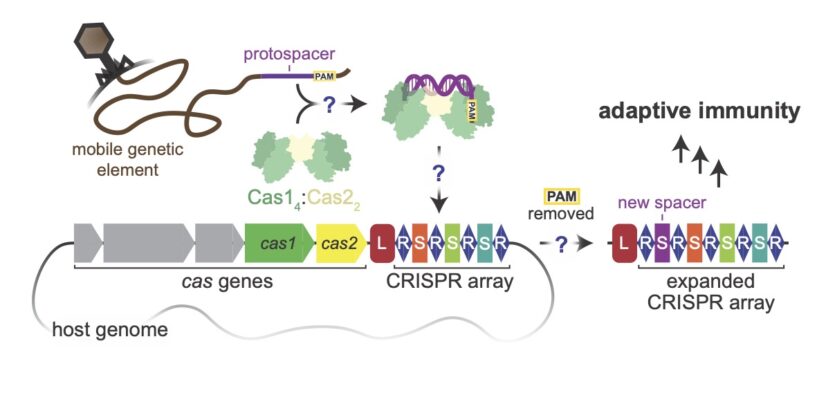
The natural Cas1–Cas2/exonuclease fusion (trimmer-integrase) catalyzes coordinated DNA capture, trimming, and integration. [Owen T. Tuck]
Tuck is fascinated by how the CRISPR array represents a miniature model of directed evolution and draws parallels between it and Lamarck’s giraffes. “Lamarck’s idea that the more a giraffe stretched its neck, the longer its baby’s neck would be was wrong,” said Tuck. “But the bacteria are kind of stretching their necks here because they’re making a targeted mutation for their selective advantage within a single generation. All their progeny now has the inserted sequence. Bugs are smart!”
Many modern biotech tools, such as CRISPR, were initially inspired by discoveries in bacterial biochemistry. Not only that, but bacteria are aiding in the advancement of new medical techniques. Tuck exclaims, “They’re amazing—absolutely amazing!”
*This post is from www.genengnews.com/topics/genome-editing/crisprs-tricks-how-cas-enzymes-capture-trim-and-integrate-foreign-dna/

