CRISPR Screens Should Be Exhaustive, Not Exhausting
Most CRISPR screens to date have been knockout screens; that is, they have relied on guide RNA–directed nucleases to introduce double-stranded breaks at targeted DNA sites. When these breaks are repaired, errors such as frameshift mutations typically result, disabling target genes. However, some CRISPR screens use “dead” nucleases that can work with transcriptional inhibitors or activators to allow inhibition (knockdown) or activation screening. [Design Cells/Getty Images]
By Ashleen Knutsen
Identifying a few key genes within the vastness of the human genome was once akin to finding the proverbial needle in a haystack. But with the introduction of functional genomic screening, scientists gained the ability to track down important genes quickly and efficiently.
Popular functional genomic screening methods include RNA interference (RNAi) and CRISPR screening. Each method presents certain advantages and disadvantages. In general, CRISPR screening—the subject of this article—is preferred if experimental goals are best pursued via complete gene knockout, as opposed to partial gene knockdown. Also, CRISPR screening is a good choice if there is no need to study essential genes. Even here, however, CRISPR screening is possible if screening procedures incorporate a catalytically “dead” nuclease, as is the case in CRISPR interference (CRISPRi) workflows.
CRISPR screening is valued for its signal strength and specificity. The method’s signal strength is attributed to the complete and permanent gene disruption that CRISPR achieves via the creation of double-strand breaks in target DNA. The method’s specificity is attributed to the relative infrequency (compared to RNAi) of off-target effects.
With CRISPR screening, scientists have a powerful tool for studying disease and potential treatments. For instance, the genes identified via CRISPR screening can be key to the mode of action of a certain compound, play a role in the resistance or sensitivity to a particular drug, or even cause a genetic disorder.
To ensure that CRISPR screening provides high-quality results, investigators must execute well-designed workflows. For example, investigators must decide between pooled, arrayed, and single-cell approaches. Also, investigators must execute suitable strategies for building libraries of nuclease-guiding, gene-targeting molecules, that is, guide RNAs (gRNAs). For details on all of these points, read the rest of this article, which includes insights from several CRISPR screening experts.
Getting started
“The choice of the cell model is key in this very first step of drug discovery to start a successful program,” says Anne-Marie Zuurmond, PhD, director, Charles River Laboratories. “The model should be representative of the disease to be mimicked.”
With CRISPR screening, cell models can be anything from standard cell lines to human primary or stem cells. Once the model is selected, the gene targets need to be chosen, which can be the entire genome or a subsection.
Next, the CRISPR modality or modalities are chosen. Most commonly, CRISPR knockout is used, but CRISPR activation and CRISPR inhibition can also be utilized, as they can provide a more detailed view of the interaction networks in gene functions.
The format of the CRISPR screen, which is how the gRNA molecules are delivered to the cells, is also selected, as well as the CRISPR nuclease and gRNA molecules themselves. Then, the delivery methods are optimized to provide an acceptable editing efficiency to obtain meaningful hits. These “hits” or gene targets are identified through next-generation sequencing, which determines what sequences are present or absent in the cells that have survived. Because each cell has only one gene that has been modified, this sequencing identifies which genetic sequences are needed for survival. This can be in normal conditions, in the presence of a certain treatment, or in a particular physiological condition.
Once targets are identified, they can be validated. Also, follow-up studies can be performed as needed.
Pros and cons of different formats
CRISPR screens can be performed either with a pooled approach, where all the cells are in large plates, or an arrayed approach, where each well in a multiwell plate contains a different target.
With a pooled format, the screen is usually designed so that most of the cells have only one target gene affected. Cells with disruptions that affect the activation of a particular gene or pathway can be selected for, and the gRNA molecules that are enriched or depleted indicate genes that are important for viability or pathway activation.
This method is cost-effective when screens sift through large panels of gene targets, such as the 19,000 protein-coding genes in the human genome.
Arrayed screens, on the other hand, require special equipment. For instance, an automated system that moves from well to well is needed since, in some cases, tens of thousands of wells are used.
“The advantage of pooled screening is that you can look at a large number of targets and identify gene targets essential for viability, drug resistance or sensitivity, or activation of phenotypic pathways without large-scale infrastructure or special equipment,” says Paul Diehl, PhD, chief operating officer, Cellecta. “You just have to be able to do cell culture and sequencing.”
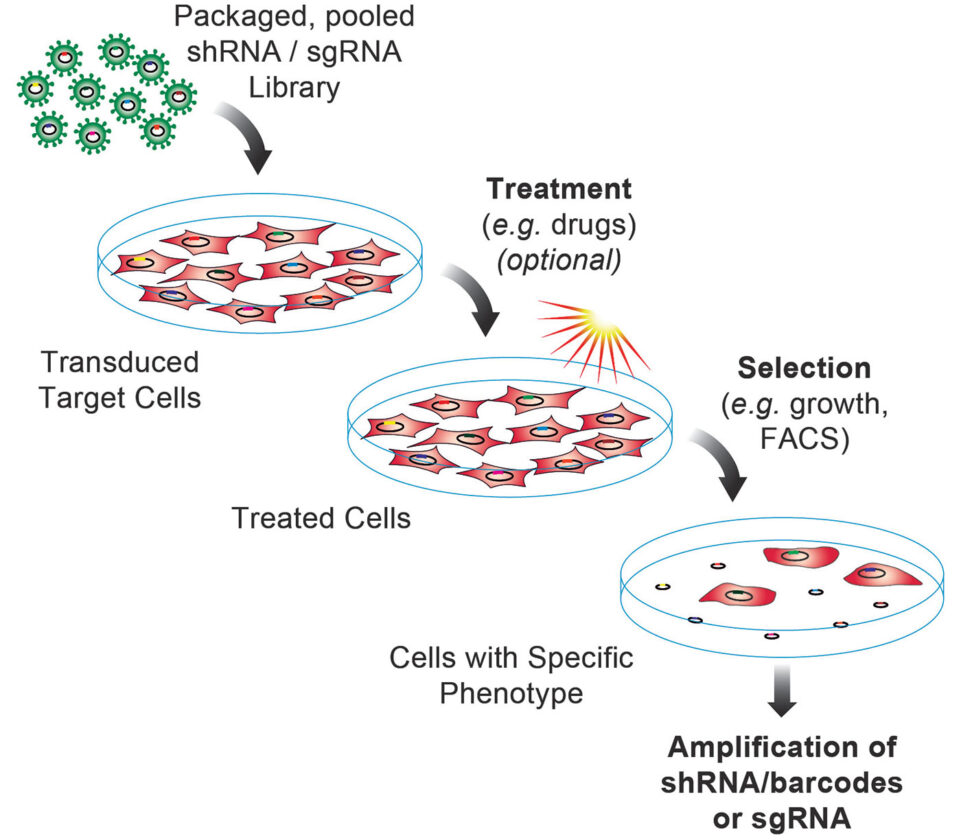
RNA (shRNA) and single guide RNA (sgRNA) libraries to help clients identify genes or genetic elements functionally regulating biological responses in cell lines or model systems of interest.
However, after sequencing, data analysis of pooled screens requires more work to see which gRNA molecules affected the phenotype. In contrast, with arrayed screens, experimenters know which gRNA was used in a particular well prior to data analysis. This allows for more sophisticated assays to be used, providing a clearer picture of the relationship between the genotype and phenotype.
Single-cell screening: The best of both worlds
A third option is single-cell CRISPR screening. With this method, a marker similar to a barcode is attached to each cell and used to facilitate tasks such as determining each cell’s expression profile.
“This approach combines the flexibility of pooled screening methodologies with the ability to examine the effect on transcription at a single-cell level,” says James Goldmeyer, PhD, associate director of product management at Revvity. (Launched last May, Revvity comprises elements that were once part of PerkinElmer. One of these elements, Horizon Discovery, retained Goldmeyer through its change in affiliation.)
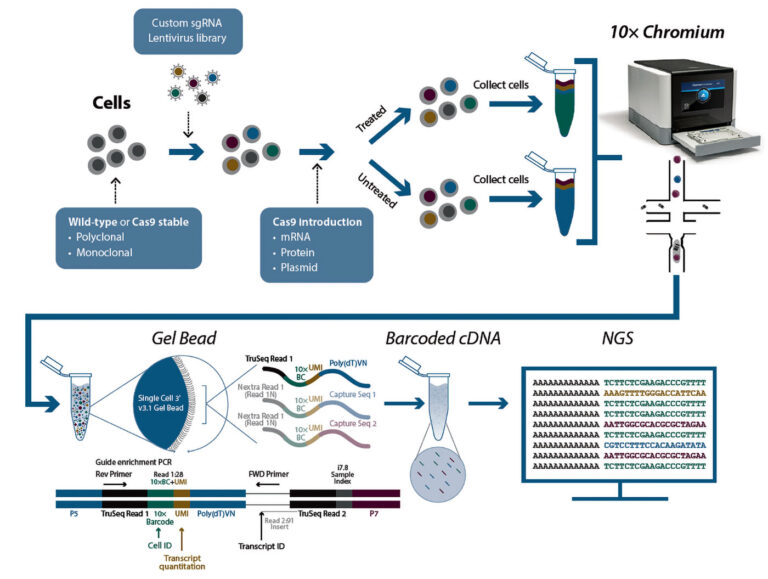
One of the services offered by Revvity is CRISPR single-cell (CRISPRsc) screening, which combines the flexibility of pooled screening with the power of transcriptomics at single-cell resolution. By interrogating the effect that a specific guide RNA has on a single-cell whole transcriptome readout, CRISPRsc screening can enable researchers to examine critical genes involved in a phenotypic outcome within a heterogenous cell population, such as a tumor or group of cells at different temporal points in a pathway
Although pooled screening links a genetic change to an observed phenotype, single-cell screening can go much further by linking specific cellular responses to a certain gRNA. According to Goldmeyer, the advantage of this approach is twofold—users can “observe nuanced changes in the transcriptome that are occurring as a direct result of the stimulus to the cell,” and they can “get a more complete picture of the level of variation in response across the cell population.”
Selecting a gRNA library
One of the most crucial steps in the CRISPR screening process is the selection of the gRNA library, the hundreds to thousands of gRNA sequences that direct the nuclease, often Cas9, to the target location within the genome.
“Design of the gRNAs used within screens has a large impact on the quality of data output from a screen,” says Mollie Schubert, product manager, Integrated DNA Technologies (IDT). “If the selected guides are not efficiently editing the locus of interest, then you may suffer from false negatives. Conversely, if guides aren’t specific and create off-target edits, then you may suffer from false positives.”
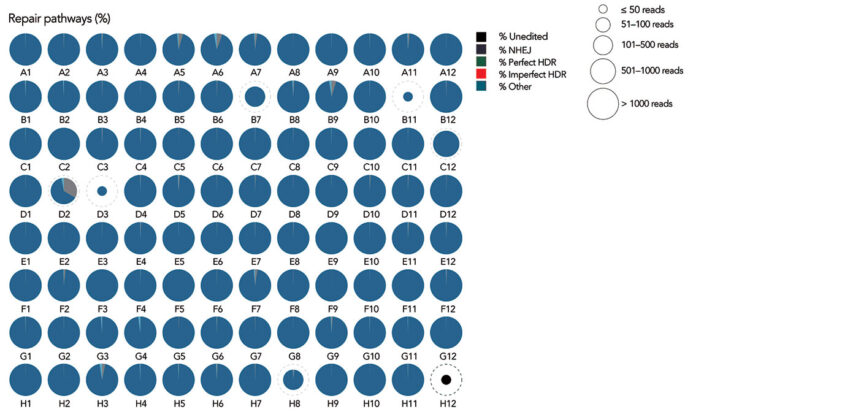
Integrated DNA Technologies (IDT) develops technologies for diverse genomics applications. For example, the company develops Alt-R Custom CRISPR gRNA Libraries. In a recent study, IDT used its technology to implement a three-guide-per-gene design and provide a high-throughput editing solution with more than 90% nonhomologous DNA end joining in almost all (90/91) targeted amplicons.
There are typically many guides that can be used for any particular gene target, and scientists often choose multiple guides to increase the likelihood of creating a functional knockout. But their levels of efficiency and selectivity may vary depending on aspects of the experiment, such as the type of cells being used, how effectively CRISPR reagents are delivered to the cells of interest, and the type of nuclease and its potential editing activity.
Genomics service providers often offer predesigned, off-the-shelf gRNA libraries, but some also can provide a custom approach where a library is created to match the customer’s needs.
For instance, IDT’s Alt-R Custom CRISPR gRNA libraries can be made for different CRISPR systems, and they are compatible with flexible options for delivery. The company has also developed a CRISPR-Cas9 library design tool that chooses gRNAs with high selectivity.
Building a better library
A custom library isn’t always necessary, however, and many meaningful experiments can be executed with a predesigned option.
In addition to its custom options, Thermo Fisher Scientific offers predesigned gRNA libraries. These libraries are built using a proprietary algorithm, and then they are packaged in lentivirus particles for delivery to cells.
This algorithm chooses gRNAs based on how potent and efficient they are. The company says that this saves researchers time by using gRNAs optimized for high activity and low off-target profile.
“The gRNAs are scored based on specificity and filter out overlapping sequences to yield up to eight gRNA designs per target,” says Jon Chesnut, PhD, senior director of cell biology R&D at Thermo Fisher. “The CRISPR gRNAs used in our libraries are highly specific to minimize or eliminate off-target effects that may mask the gene-specific silencing results.”
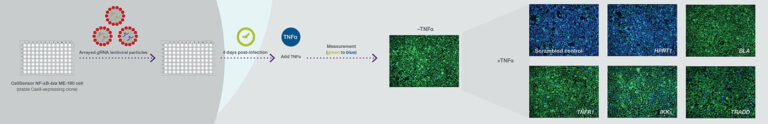
After introducing a LentiArray library of gRNA lentiviral particles to its Invitrogen CellSensor cell line, Thermo Fisher Scientific stimulated the cells with TNFα. In unedited cells, the ratio of green to blue fluorescence decreased. In cells infected with lentiviral particles (which carried gRNA that effectively disrupted the NF-κB pathway), there was a high ratio of green to blue fluorescence. Complete knockout of TNFR1 was observed in nearly 100% of the cells.
One customer used a LentiArray CRISPR library to research pediatric brain tumors and identified a novel target and potential treatment strategy, the company says. But this is just the tip of the iceberg for uses for functional genomic screening, which is becoming ever more efficient thanks to newer and more precise gene editing technologies.
For instance, some companies, such as Revvity, are offering base editing. “Base editing offers the ability to precisely edit a specific target base using a deaminase, rather than relying on the cell’s own DNA repair mechanisms,” Goldmeyer notes. “This base editing precision is ideal when you know exactly which base change you are interested in.”
But like every other step in the screening process, selecting the method that best suits an experiment and a customer’s needs—whether it’s RNAi, CRISPR, or even base editing—depends on the study design, the desired testing outcome, timeline, and budget.
*This post is from www.genengnews.com/topics/genome-editing/crispr-screens-should-be-exhaustive-not-exhausting/

