[Literature Review] CRISPR Detection Novel Breakthrough: Harnessing CrisprAIE for Accurate and Rapid Detection

Rapid and accurate detection of nucleic acid biomarkers is essential for diagnosing and monitoring infectious diseases. Traditional methods like qPCR and sequencing are effective but are limited by their complexity, cost, and time requirements. Although isothermal nucleic acid amplification simplifies the process, it often results in nonspecific amplification and false positives.
The CRISPR/Cas system, a natural immune mechanism in microorganisms, enhances sensitivity and specificity in nucleic acid detection by targeting specific sequences and enabling precise collateral cleavage of non-targeted single-stranded nucleic acids. CRISPR-based diagnostic methods (Crispr-Dx), utilizing Cas12 or Cas13, detect various targets such as viruses and genetic mutations through the cleavage of single-stranded DNA or RNA.
However, challenges remain. Signal generation in Crispr-Dx assays heavily relies on the trans-cleavage of single-stranded nucleic acids, which limits signal efficiency, sensitivity, and stability, especially in complex biological environments.

Original Article Link:https://doi.org/10.1038/s41467-024-52931-0
To address this issue, Chinese researchers have introduced CrisprAIE, a novel system that integrates CRISPR/Cas reactions with "one to more" aggregation-induced emission luminogens (AIEgens). It enhances fluorescence through the trans-cleavage of Cas proteins linked to AIEgen-incorporated double-stranded DNA. The findings were published in Nature Communications under the title "CRISPR/Cas-mediated 'one to more' lighting-up nucleic acid detection using aggregation-induced emission luminogens".
CrisprAIE demonstrates exceptional performance in clinically detecting norovirus and SARS-CoV-2 without relying on amplification methods. Its diagnostic capabilities are further enhanced by incorporating spherical nucleic acid-modified AIEgens and a portable cellphone-based readout device. This combination significantly improves sensitivity compared to traditional CRISPR-based diagnostics. Overall, CrisprAIE is positioned as a versatile signal generation strategy that can greatly boost the detection efficiency of existing CRISPR diagnostics.
I. Crispr-Driven AIEgen Lighting-Up Readout
Cationic AIEgens interact with DNA strands through electrostatic interactions, activating restricted intramolecular motion (RIM) and resulting in enhanced fluorescence. TPBT, a typical cationic AIEgen with four positively charged pyridine groups, provides a label-free method for detecting double-stranded DNA (dsDNA) and single-nucleotide polymorphisms. It also features a large Stokes shift and greater resistance to photobleaching compared to traditional fluorophores like FAM.
Recognizing that Cas nucleases prefer single-stranded oligonucleotides, researchers developed ssDNA- and dsDNA-modified AIEgen reporters. The dsDNA-based probes produced stronger fluorescent signals by more effectively restricting AIEgen motion. The CrisprAIE system, which combines CRISPR/Cas reactions with DNA-modified AIEgen reporters, was created for nucleic acid detection.
This system demonstrated enhanced sensitivity and signal output, particularly with Q-dsDNA/AIEgens and Q-dsDNA/AIEgens-Q reporters, achieving a significantly lower limit of detection (LOD) than conventional methods. When integrated into the CrisprAIE system, these optimized reporters reduced the LOD by approximately 80-fold. This improvement was due to more effective restriction of AIEgens in dsDNA, efficient signal amplification, and a higher signal-to-noise ratio.
Additionally, CrisprAIE showed excellent stability under cryogenic conditions, compatibility with lyophilized CRISPR reagents, and adaptability with other AIEgens. These features make CrisprAIE a promising tool for high-sensitivity nucleic acid detection in complex biological systems.
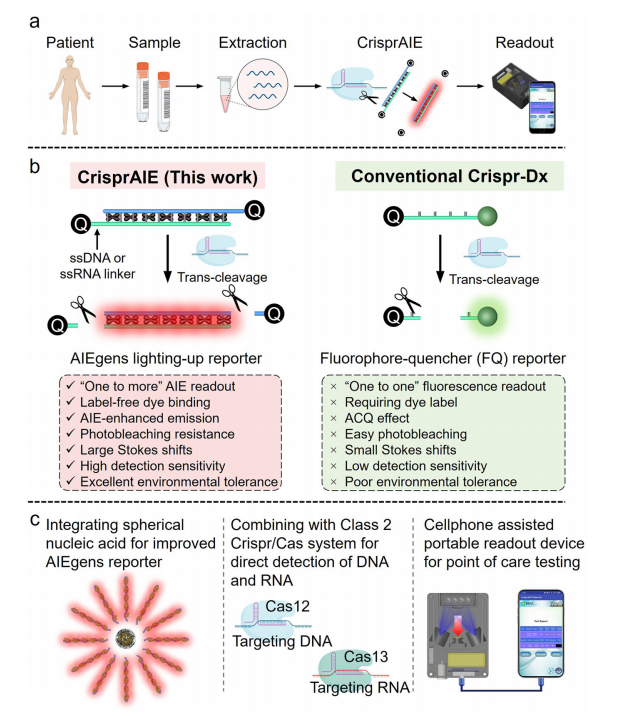
Fig.1 Schematic of the developed CrisprAIE assay
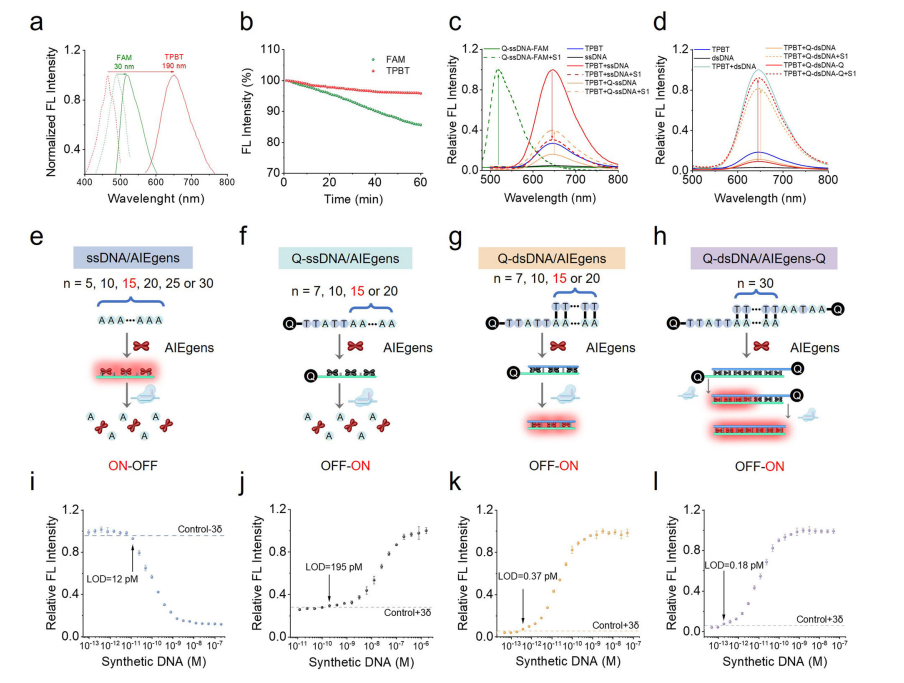
Fig.2 Construction of a Crispr-driven AIEgen lighting-up readout
II. RNA Quantification Using CrisprAIE Combined with RT-RPA
The CrisprAIE system, combined with RT-RPA (reverse transcription-recombinase polymerase amplification), has been developed for highly sensitive and specific RNA detection. This system is especially useful for detecting viral RNA in clinical samples. It integrates Q-dsDNA/AIEgen-Q fluorescence reporting with Cas12-based RNA detection, enabling a single-tube isothermal assay that combines reverse transcription and amplification. When target RNA is present, it activates Cas12a, which cleaves and triggers fluorescence, enabling rapid and accurate detection.
For norovirus detection, the CrisprAIE system accurately identified viral RNA in clinical vomitus samples, with results consistent with RT-qPCR. It demonstrated a lower limit of detection (LOD) and higher sensitivity compared to traditional methods, confirming its efficiency and reliability.
The versatility of the system was further showcased by quantifying SARS-CoV-2 RNA, where the CrisprAIE method achieved an AUC of 0.969, highlighting its potential as a powerful tool for RNA quantification in clinical diagnostics.
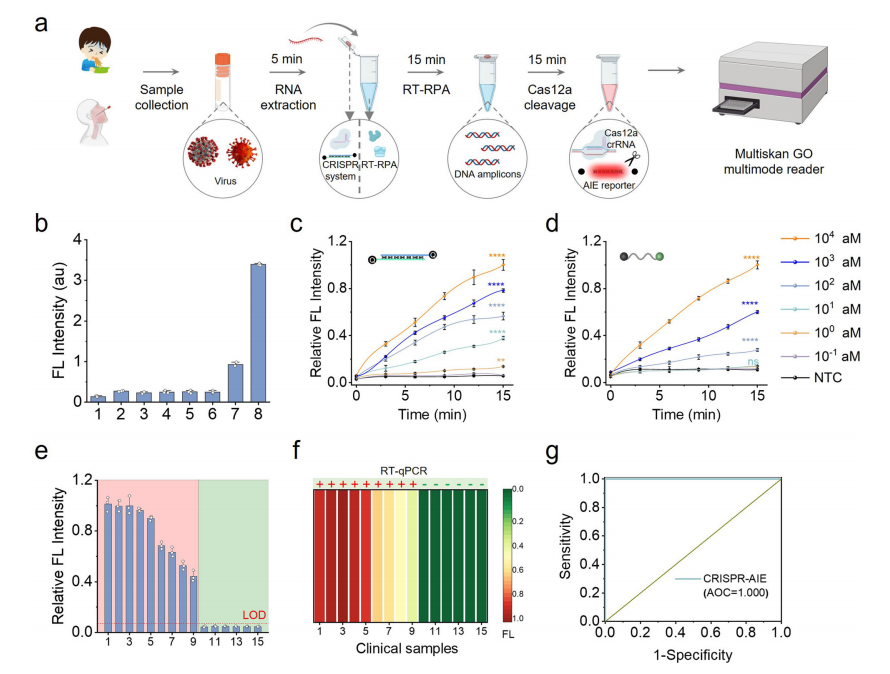
Fig.3 Combination of RT-RPA and CrisprAIE detects norovirus RNA up to attomolar concentration
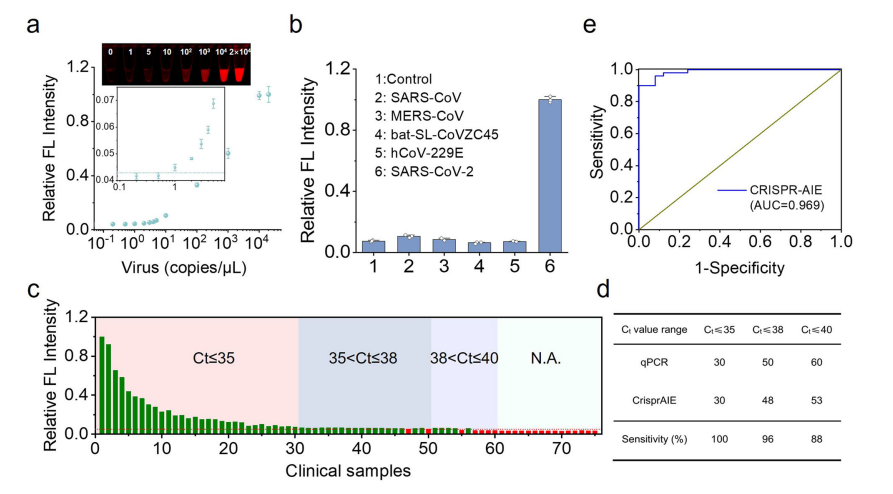
Fig.4 Combination of RT-RPA and CrisprAIE detects SARS-CoV-2 RNA to single copy level
III. Improved Sensitivity and Stability of CrisprAIE Using AIEgen-Assisted SNA Reporters
The sensitivity and stability of the CrisprAIE system were enhanced by using AIEgen-assisted SNA (Spherical Nucleic Acid) reporters. Traditional linear DNA reporters, while effective, were vulnerable to nuclease degradation in biological fluids, leading to false positives. In contrast, SNA/AIEgen reporters combined nanoparticles with nucleic acid probes, providing greater stability, enhanced fluorescence quenching, and increased resistance to nucleases.
The developed SNA/AIEgen reporter achieved a limit of detection (LOD) as low as 55.7 fM, significantly surpassing traditional Q-dsDNA/AIEgen-Q and Q-ssDNA-FAM reporters. It maintained stability and accuracy even in complex biological samples, such as serum. The SNA/AIEgen-based CrisprAIE system effectively detected various clinically relevant targets, including norovirus, SARS-CoV-2, and HPV, with minimal matrix interference, demonstrating its robustness in field diagnostics.
When combined with RT-RPA, the SNA@AIEgen reporters further improved the detection sensitivity of SARS-CoV-2 to 0.1 copies/µL, a tenfold increase over previous methods. The system also exhibited reproducibility and consistency across multiple batches, underscoring its reliability for real-world viral diagnostics.
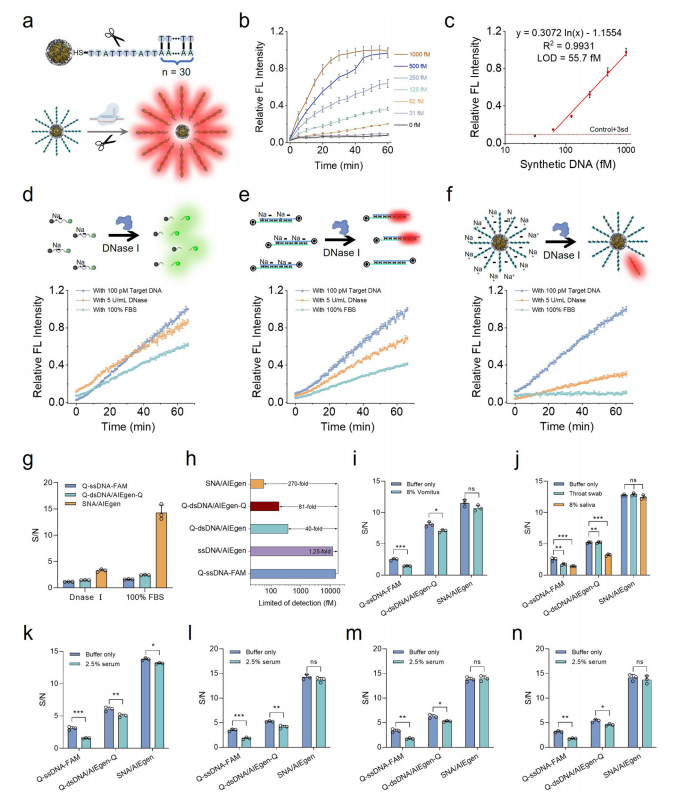
Fig.5 AIEgen-assisted SNA reporters enhance the CrisprAIE detection of target DNA in complex samples
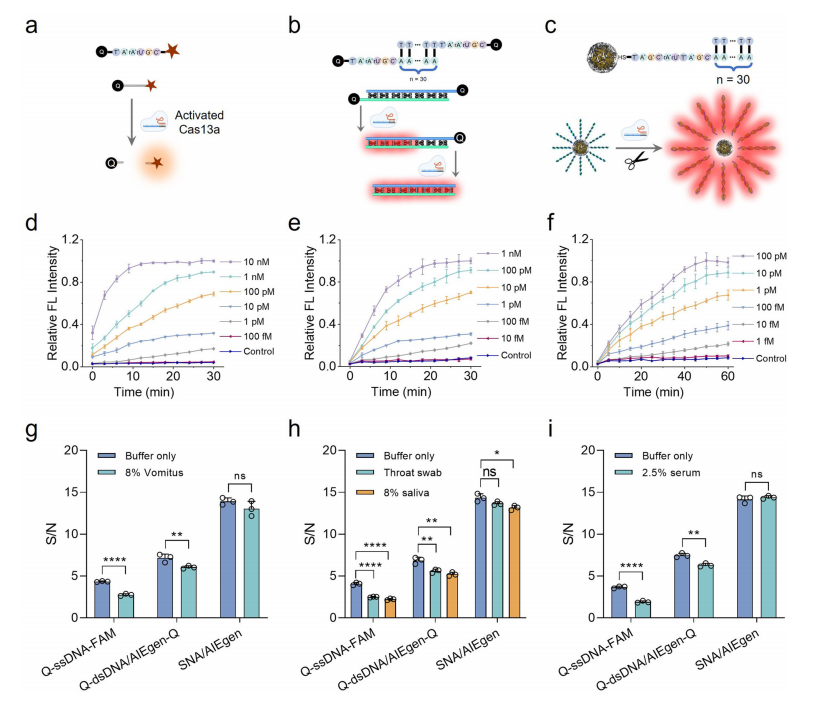
Fig.6 Expanding CrisprAIE for the direct detection of target RNA in complex samples
IV. Expanding CrisprAIE for the Direct Detection of Target RNA
The CrisprAIE system was expanded to allow direct detection of RNA targets using Cas13a. Unlike Cas12a, which requires reverse transcription of RNA into DNA, Cas13a can directly recognize RNA. The Cas13a-based CrisprAIE utilized an ssRNA linker in the Q-dsDNA/AIEgen-Q and SNA/AIEgen reporters, enabling effective RNA detection through Cas13a's trans-cleavage of ssRNA.
The CrisprAIE system achieved 10- to 100-fold greater sensitivity compared to conventional Q-ssRNA-FAM reporters, depending on the reporter used. It successfully detected synthetic RNA targets, including norovirus, SARS-CoV-2, and Ebola virus, even in complex biological samples such as vomitus, throat swabs, saliva, and serum. The SNA/AIEgen reporter demonstrated superior resistance to matrix interference, showcasing its robustness and potential for field diagnostics of RNA targets.
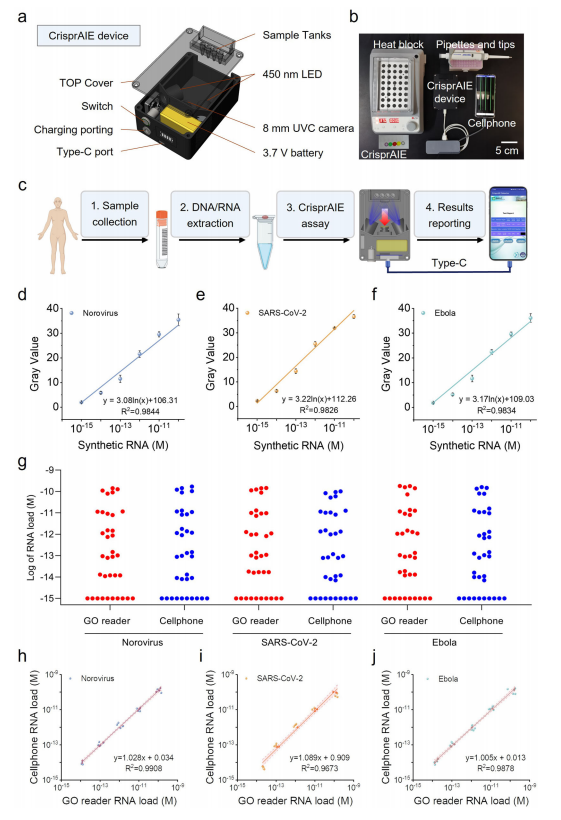
Fig.7 Portable device adapting CrisprAIE for POC testing
V. Adapting CrisprAIE for POC Testing
CrisprAIE technology significantly enhances detection sensitivity compared to traditional Crispr-Dx, while maintaining a straightforward testing process, making it ideal for point-of-care (POC) applications. A portable, cellphone-based CrisprAIE assay platform has been developed, featuring a 3D-printed body, LED illuminators, and an electrical circuit to capture fluorescent images.
The testing process involves adding a sample to a tube containing RPA and CrisprAIE reagents, incubating for 30 minutes, and displaying the results on a cellphone. The cellphone-based CrisprAIE assay demonstrated analytical performance comparable to a commercial Multiskan GO multimode reader, with similar detection sensitivity and dynamic range for synthetic RNA targets. Correlation analysis further confirmed the reliability of this assay for POC diagnostics.
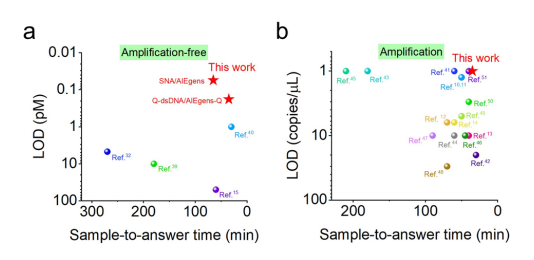
Fig.8 Comparison between CrisprAIE and reported Crispr-Dx with or without amplification
Discussion
The emergence of infectious diseases like COVID-19 underscores the urgent need for rapid and reliable diagnostic technologies that can be deployed in the field. The CrisprAIE assay combines DNA-enhanced AIEgen lighting, offering improved sensitivity and stability over traditional methods. It can detect DNA and RNA at femtomolar concentrations without amplification and achieve attomolar sensitivity with isothermal amplification.
The integration of DNA-enhanced AIEgen lighting significantly boosts the sensitivity of Crispr/Cas-based detection. A portable, cellphone-assisted CrisprAIE device demonstrates performance comparable to commercial fluorescence readers. While AIEgen probes offer advantages such as high signal-to-noise ratios and excellent photostability, they also face certain limitations. These include susceptibility to interference from complex samples and the requirement for nucleic acid extraction. Despite these challenges, ongoing research is focused on optimizing AIEgen probes for broader diagnostic applications and enhancing their deployability in field settings.
In conclusion, this work introduces the first AIEgen-enhanced Crispr-Dx assay for sensitive and accurate nucleic acid detection, showing significant potential for diagnosing infectious diseases. Future improvements for CrisprAIE could leverage advances in Cas nuclease evolution, crRNA structure optimization, and amplification primer screening. Moreover, enhancing signal reporter systems with high quantum yield AIEgens and incorporating digital reading devices could further improve its performance.
EDITGENE boasts its own independently developed protein purification platform.Our Cas enzymes have demonstrated significantly superior activity compared to other market enzymes through rigorous testing, consistently achieving the highest standards in purity, activity, and sensitivity. Be on the lookout for our isothermal amplification kit and CRISPR detection kits, efficient with minimal equipment requirements and short reaction times—Coming soon!
Recent Blogs:
1.[Weekly News] Breakthrough gRNA Algorithm Supercharges CRISPR Detection Sensitivity
2.[Literature Review] CRISPR Screening Reveals Aging Regulators in Neural Stem Cells
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






