[Cutting Edge] Gene Editing Breakthrough: PKM1, PBRM1, and S100A9 Knockout Cells Reveal New Cancer Treatment Clues

Gene knockout cells refer to cells that have undergone gene editing using CRISPR technology. By designing specific sgRNA to target the cleavage site of the desired gene, the Cas9 protein binds with the sgRNA to “cut out” the target gene from the cell, thereby achieving gene knockout. Gene knockout cells have broad applications in fields such as life sciences, medicine, and drug development, including the creation of disease models, drug screening, target validation, and gene therapy by knocking out pathogenic genes to treat diseases.
This article reviews three studies on gene knockout cells to provide insights into the latest developments in this cutting-edge technology.
I. CRISPR/Cas9 System Successfully Knocks Out PKM1 Gene, Optimizing Lactate Production in CHO Cells
Original Article Link: https://doi.org/10.1002/biot.201800332
Chinese hamster ovary (CHO) cells are one of the most commonly used mammalian cell lines in the biopharmaceutical industry, widely applied in producing various therapeutic proteins such as antibodies, hormones, and other recombinant proteins.
Pyruvate kinase muscle (PKM) is an enzyme responsible for converting PEP to pyruvate in glycolysis, where pyruvate can be used for energy production or converted into lactate. In CHO cells, the expression level of PKM1 is directly associated with lactate production.
By using CRISPR/Cas9 technology to knock out the PKM1 gene, researchers were able to eliminate lactate accumulation. In this study, researchers used the CRISPR/Cas9 system to knock out the PKM1 gene in CHO cell lines to investigate its role in lactate production and to validate the direct relationship between PKM1 expression and lactate accumulation.
The results showed that in the PKM1-knockout cell lines, no lactate accumulation was observed, even under conditions that typically promote lactate production. This demonstrates that PKM1 activity is essential for lactate generation. Furthermore, the PKM1-knockout cell lines maintained similar productivity and viability compared to wild-type cells, indicating that PKM1 deletion does not negatively impact cell survival or production capacity.
In industrial production, high levels of lactate production in CHO cells can lead to reduced cell viability and productivity, negatively affecting the efficiency of the manufacturing process and the quality of the final product. Lactate accumulation can also cause a decrease in the culture medium's pH, requiring the addition of alkaline substances to regulate the pH, which can increase osmotic pressure and further hinder cell growth and protein production.
Gene editing techniques such as CRISPR/Cas9 can knock out or suppress PKM1 expression, helping to reduce or eliminate lactate production during CHO cell cultivation, thereby improving production efficiency and product quality.
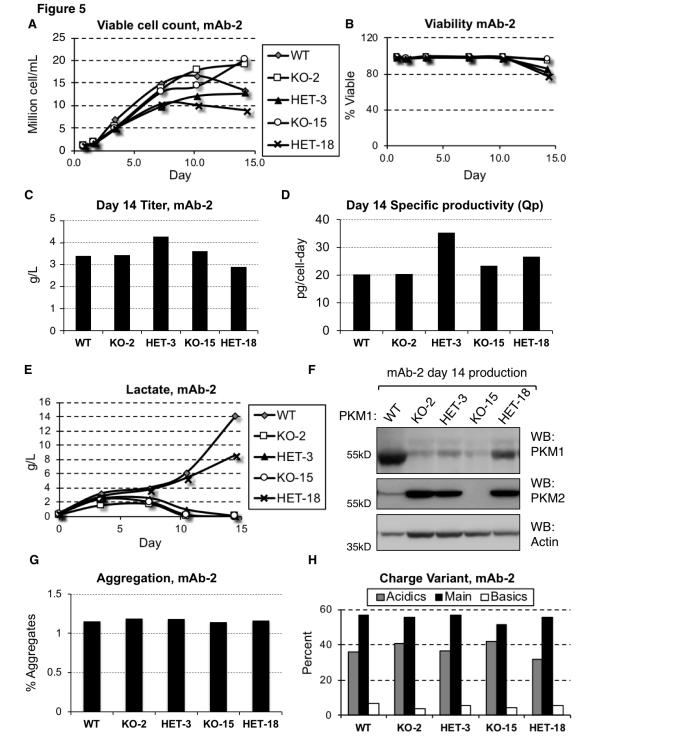
Figure 1: Reducing or Eliminating PKM1 Expression Prevents Lactate Production in mAb-2 Cell Lines
II. PBRM1 Gene Knockout Linked to Lower Immunogenic TME in ccRCC
Original Article Link: https://doi.org/10.1038/s41467-020-15959-6
Renal cell carcinoma (RCC) is one of the top ten most common malignancies worldwide, with clear cell RCC (ccRCC) being the most prevalent histological subtype. ccRCC frequently harbors secondary gene mutations, including mutations in the polybromo-1 (PBRM1) gene.
PBRM1 is part of the SWI/SNF chromatin remodeling complex, and approximately 40% of ccRCC cases involve PBRM1 mutations. To date, the data regarding the effect of PBRM1 loss on immune responses have been inconsistent, prompting researchers to investigate how PBRM1 loss affects the tumor microenvironment (TME) in ccRCC.
In this study, researchers used CRISPR/Cas9 gene editing technology to knock out the Pbrm1 gene, generating Pbrm1-knockout Renca mouse RCC cell lines and derived tumors. Using the Renca-BALB/c immune mouse model system, they assessed immune markers in different mouse tumors via immunohistochemistry (IHC), focusing on CD3, CD8, CD4, PD-1, and P-STAT1.
The results revealed that in control group tumors, the levels of CD3, CD4, CD8 T cells, and p-STAT1-positive cells were significantly higher compared to Pbrm1-knockout mice, and the expression of immune checkpoint protein PD-1 was also higher in the control tumors. The Pbrm1-knockout cell lines used to construct tumor models demonstrated that Pbrm1 deficiency is associated with a lower immunogenic tumor microenvironment, which has important implications for cancer immunotherapy.
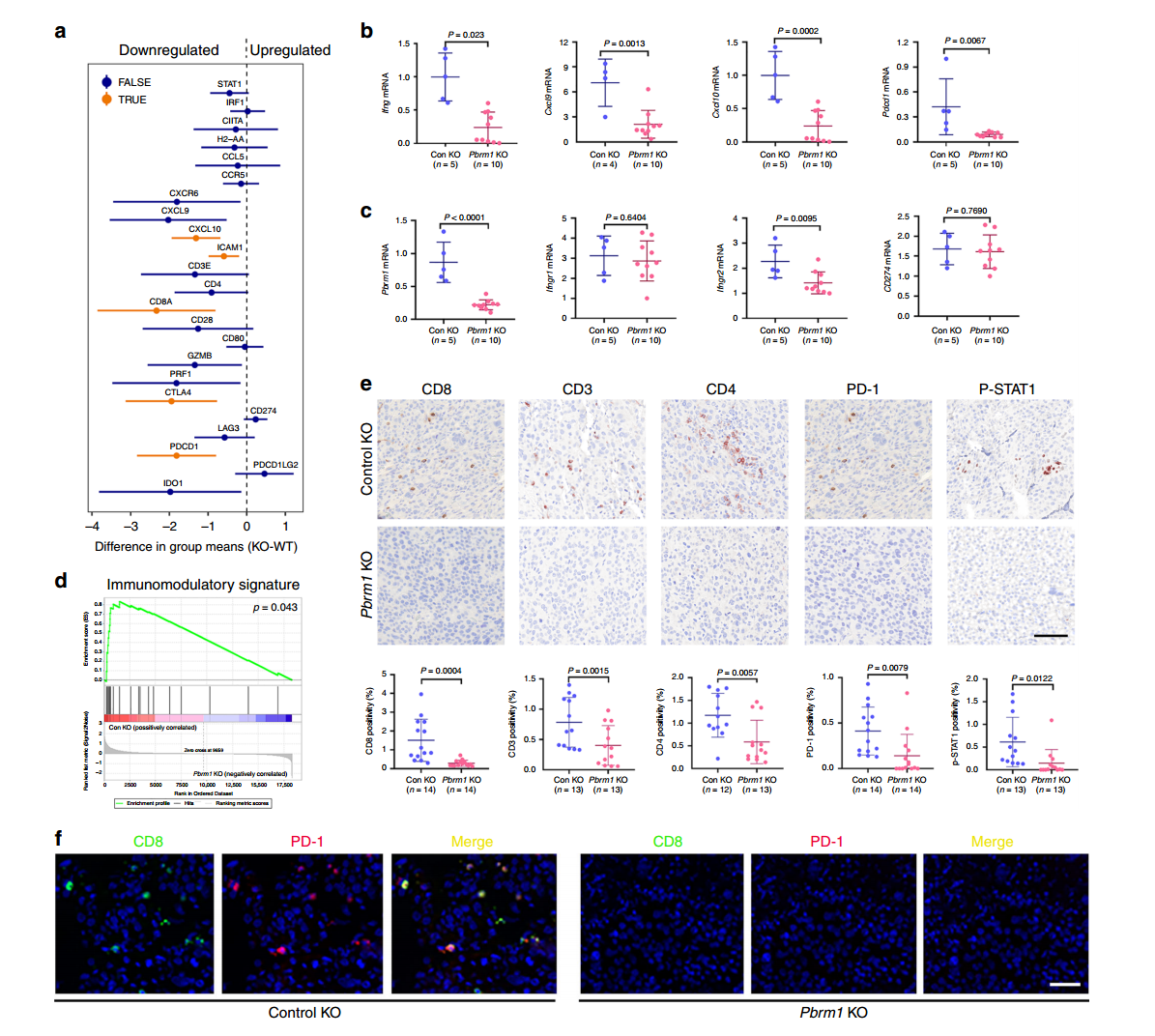
Figure 2: Pbrm1 Knockout Tumors Are Associated with a Low Immunogenic TME
III. S100A9 Knockout Offers Further Insight into the Pathogenesis of SICM
Original Article Link: https://doi.org/10.2147/JIR.S457340
Sepsis-induced cardiomyopathy (SICM) is a common form of heart dysfunction caused by sepsis, with a complex pathophysiology involving several key factors such as myocardial suppression, mitochondrial dysfunction, inflammatory responses, and oxidative stress.
S100A9 (S100 calcium-binding protein A9) is a calcium- and zinc-binding protein that plays a critical regulatory role in inflammation and immune responses. However, the exact mechanisms by which S100A9 induces septic heart failure and myocardial injury remain poorly understood.
Researchers used CRISPR-Cas9 technology to knock out the S100a9 gene in mice and exposed primary cardiomyocytes to lipopolysaccharide (LPS) to simulate SICM. By measuring inflammatory markers and S100A8/9 expression, they successfully modeled sepsis-induced cardiomyopathy. Echocardiographic results revealed that S100a9 gene knockout alleviated sepsis-induced cardiomyopathy and reversed myocardial damage compared to wild-type controls.
Transmission electron microscopy (TEM) analysis of mitochondrial cristae density showed that the wild-type control group had uniformly structured mitochondria, while the septic wild-type group exhibited significantly increased heterogeneity in mitochondrial size and number. Knocking out S100a9 in mice reduced mitochondrial damage and restored normal structure.
This study demonstrates that S100A9 is involved in sepsis-induced cardiac injury and dysfunction, revealing potential mechanisms through which S100A9 induces oxidative stress and mitochondrial dysfunction. The use of CRISPR-Cas9 technology to knock out S100A9 gene in cells provides a valuable model for studying the gene’s role in SICM, offering potential therapeutic insights and contributing to the understanding of SICM pathogenesis.
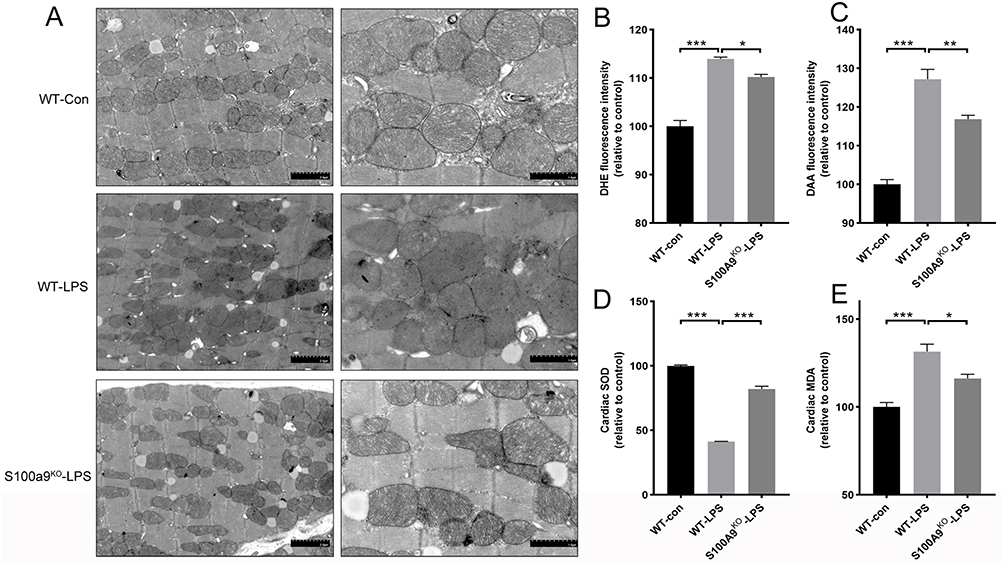
Figure 3: S100a9 gene knockout alleviates mitochondrial damage and oxidative stress
EDITGENE offers over 3,800 knockout cell lines in stock, including those mentioned in the articles above such as PKM1 , PBRM1 , and S100A9 . We provide one-stop scientific research support, with orders delivered within a week!
Recent Blogs:
1.[Literature Review] CRISPR Knockout Provides Novel Insight into CENP-E-mediated Cell Division
2.[Weekly News] CRISPR screening reveals new mechanism: IL-4 contributes to CD8+ CART cell failure
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






