[Literature Review] CRISPR Knockout Provides Novel Insight into CENP-E-mediated Cell Division

Accurate alignment and segregation of chromosomes are crucial for maintaining chromosomal stability in metazoans. Mitotic kinesins such as Centromere-associated protein E (CENP-E) play a pivotal role in regulating microtubule dynamics, spindle organization, and chromosome alignment. As a kinesin-7 motor protein, CENP-E accumulates during the late G₂ phase and is essential for kinetochore-microtubule attachment and the regulation of the spindle assembly checkpoint during mitosis.
Depletion of CENP-E through various methods results in chromosome misalignment and activation of the mitotic checkpoint, while complete knockout leads to cell cycle arrest and even cell death. In mice, homozygous deletion causes early developmental arrest, whereas heterozygous deletion leads to chromosome missegregation and tumor development.
To coordinate chromosome alignment and segregation, CENP-E, along with other proteins like Dynein and Aurora B, acts as a linker between chromosomes and kinetochore fibers. BubR1 protein recruits CENP-E for checkpoint activation; however, the impact of CENP-E deletion on spindle checkpoint activation still remains uncertain.
To address this situation, researchers from China employed CRISPR-Cas9 gene editing technology, alongside high-throughput screening, chemical inhibition, and advanced microscopy techniques, to investigate the critical role of CENP-E in chromosome alignment and stability. Their findings were published in the journal Cell Proliferation under the title "Kinesin-7 CENP-E mediates centrosome organization and spindle assembly to regulate chromosome alignment and genome stability". The results indicated that both CENP-E inhibition and deletion resulted in chromosome misalignment, spindle disorganization, and defective chromosome segregation.
Additionally, CRISPR-Cas9 knockout of CENP-E caused significant defects in chromosome congression and alignment, which triggered activation of the spindle assembly checkpoint and subsequent cell cycle arrest. These findings contribute to a more comprehensive understanding of the molecular functions of CENP-E in maintaining chromosome alignment and regulating cell cycle progression.

Original Article Link: https://doi.org/10.1111/cpr.13745
I. CENP-E inhibition results in chromosome misalignment, mislocalization, and multipolar spindle formation
CENP-E proteins are highly expressed during the G₂/M phase, first localizing to kinetochores in prometaphase and metaphase, and later moving to the central spindle during anaphase and cytokinesis. To study the role of CENP-E in HeLa cells, researchers used the specific inhibitor GSK923295 to block its ATPase activity, effectively immobilizing CENP-E on microtubules.
The results showed that inhibiting CENP-E caused significant chromosome misalignment at spindle poles, shortened spindle length, and increased spindle width. The degree of chromosome disorganization was dose-dependent, with higher concentrations of GSK923295 leading to more severe effects. Fluorescence analysis further revealed an increased accumulation of CENP-E at the kinetochores of misaligned chromosomes near the spindle poles, while its localization at the kinetochores of aligned chromosomes diminished.
CENP-E inhibition also disrupted the cell cycle in HeLa cells, leading to a decrease in interphase cells and an increase in metaphase cells, along with a reduction in anaphase cells. Additionally, inhibition resulted in a higher frequency of bipolar and multipolar spindle formation, indicating defects in spindle assembly. Measurements of γ-tubulin areas and fluorescence intensity showed that CENP-E inhibition caused centrosome dispersal and reduced γ-tubulin concentration at spindle poles, further disrupting spindle organization. These findings underscore the critical role of CENP-E in chromosome alignment, spindle assembly, and overall cell division architecture.
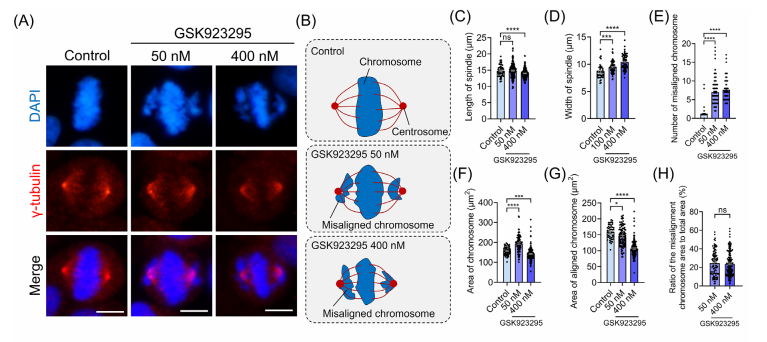
Fig.1 CENP-E inhibition resulted in chromosome misalignment and the abnormal localization of CENP-E proteins at metaphase
II. CENP-E deletion leads to chromosome misalignment and centrosome disorganization
To explore the specific functions of CENP-E in cell division, CRISPR-Cas9 gene editing was used to generate CENP-E knockout Hela cell lines. After the transfection of HeLa cell lines with CENP-E-specific sgRNA, the researchers induced double-strand breaks at the CENP-E locus, and triggered endogenous DNA repair through nonhomologous end joining, which resulted in random insertions, deletions, and substitutions.
To conduct the experiments, CENP-E+/- heterozygous and CENP-E-/- knockout clones were generated. In the knockout clones, complete loss of CENP-E protein led to significant chromosome misalignment, with chromosomes abnormally concentrated near spindle poles. In contrast, the heterozygous CENP-E+/- cells exhibited more dispersed chromosome distribution. Statistical analysis confirmed that CENP-E deletion resulted in a higher extent of chromosome misalignment and scattered chromosomal organization around the spindle poles.
Through the investigation in CRISPR-knockout CENP-E-/- cell lines, the researchers also identified the role of CENP-E in cell division during prometaphase and metaphase. CENP-E proteins, which typically accumulate at kinetochores and spindle microtubules during prometaphase, gradually disassociated from aligned kinetochores at metaphase, but remained misaligned following CENP-E inhibition or deletion.
In CENP-E-/- cells (knockout clones), complete knockout led to the formation of multipolar spindles, increased distances between spindle poles, and disrupted centrosome organization, including dispersion of y-tubulin-labeled centrosomes and reduced k-fibre width. Immunofluorescence analysis further demonstrated altered spindle pole dynamics, highlighting CENP-E's critical role in maintaining spindle pole organization, centrosome integrity, and k-fibre assembly during metaphase.
CENP-E deletion leads to chromosome misalignment and centrosome disorganization.
To explore the specific functions of CENP-E in cell division, CRISPR-Cas9 gene editing was used to generate CENP-E knockout HeLa cell lines. After transfecting HeLa cells with CENP-E-specific sgRNA, the researchers induced double-strand breaks at the CENP-E locus, triggering endogenous DNA repair through nonhomologous end joining (NHEJ), which resulted in random insertions, deletions, and substitutions.
For the experiments, both CENP-E+/- heterozygous and CENP-E-/- knockout clones were created. In the knockout clones, the complete loss of CENP-E protein led to significant chromosome misalignment, with chromosomes abnormally clustering near the spindle poles. In contrast, the heterozygous CENP-E+/- cells showed a more dispersed chromosome distribution. Statistical analysis confirmed that CENP-E deletion caused a higher degree of chromosome misalignment and disorganized chromosome distribution around the spindle poles.
Further investigation into the CRISPR-knockout CENP-E-/- cell lines revealed the role of CENP-E during prometaphase and metaphase in cell division. Normally, CENP-E proteins accumulate at kinetochores and spindle microtubules during prometaphase, but they disassociate from aligned kinetochores at metaphase.
However, with CENP-E inhibition or deletion, these proteins remained on the misaligned chromosomes. In CENP-E-/- cells, the complete knockout led to the formation of multipolar spindles, increased spindle pole distances, and disrupted centrosome organization, including dispersion of γ-tubulin-labeled centrosomes and a reduction in k-fibre width. Immunofluorescence analysis further demonstrated altered spindle pole dynamics, highlighting CENP-E's critical role in maintaining spindle pole organization, centrosome integrity, and k-fibre assembly during metaphase.
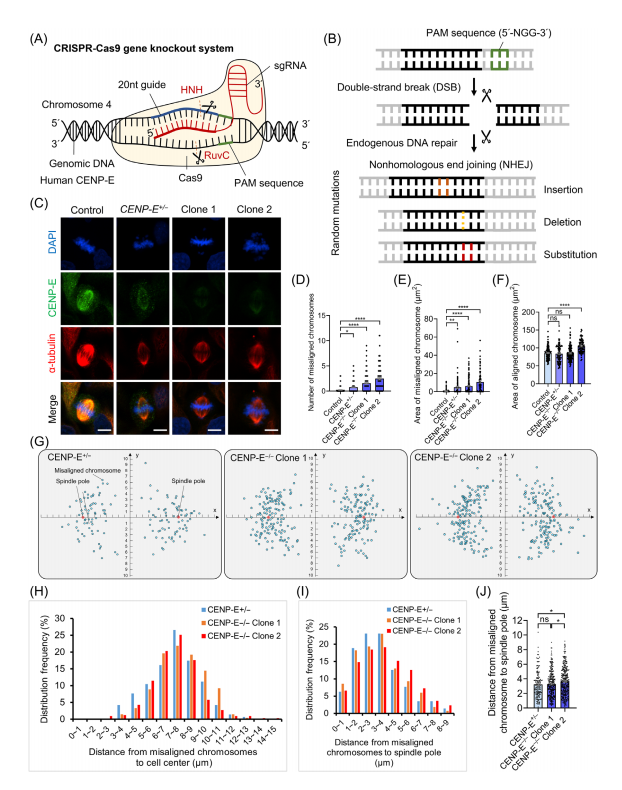
Fig.2 Construction of CENP-E knockout Hela cells using the CRISPR-Cas9 gene editing technology
III. CENP-E is essential for spindle organization, chromosome alignment and cell cycle progression in vivo
In this study, the researchers found that deletion of CENP-E resulted in the accumulation of KIF2C proteins on misaligned kinetochores, particularly around the spindle poles. In addition, the deletion also led to the decreased localization of BubR1 proteins on aligned kinetochores, while increased on misaligned ones during metaphase. Mad1 proteins were found to accumulate on misaligned kinetochores in both CENP-E+/- and CENP-E+/+ cells, indicating the activation of the Mad1-related spindle assembly checkpoint. The researchers used CENP-E knockout heterozygous mice to conduct in vivo experiments, the results showed that CENP-E deletion increased apoptosis in lymphocytes, as evidenced by the rise in TUNEL-positive and pHH3-positive cells in the spleen, without affecting spleen weight. Additionally, CENP-E deletion resulted in chromosome misalignment and mitotic spindle disorganization, leading to an increase in dividing lymphocytes and apoptotic cells.
The researchers also conducted cell colony formation assay, the results revealed that CENP-E deletion significantly decreased the number and diameter of cell colonies in HeLa cells, indicating suppressed cell proliferation. Compared to the control, analysis showed a reduction in the number of cells per clone in both CENP-E+/- and CENP-E-/- HeLa cells, while the area of interphase and metaphase cells increased in CENP-E-/-HeLa cells. Cell cycle analysis demonstrated a significant rise in G2/M phase cells after CENP-E deletion, with haploinsufficiency leading to a mild increase in these cells as well. Additionally, cell apoptosis analysis indicated an increase in both early and late apoptotic cells after CENP-E deletion. Collectively, these findings suggested that CENP-E deficiency results in cell cycle arrest, inhibited cell growth, and also increased apoptosis in cultured cells.
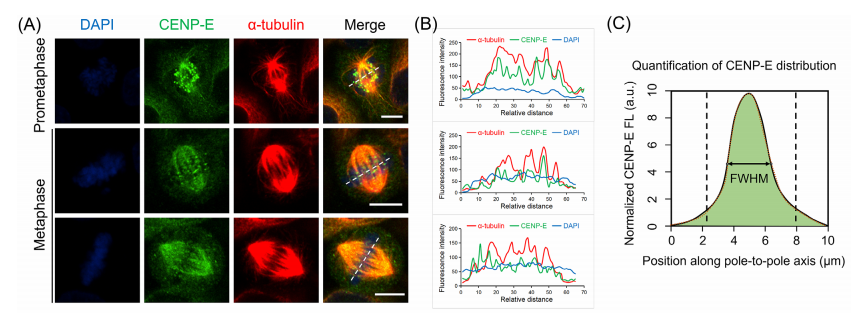
Fig.3 Dynamic localization patterns of CENP-E proteins in wild-type, GSK923295 inhibited and CENP-E knockout cell
IV. CENP-E is required for cell proliferation, cell cycle control, and chromosome stability
Through staining HeLa cell lines, the researchers investigated the effects of chromosome misalignment following CENP-E deletion. The results revealed a translocation of Aurora B proteins on the spindle in CENP-E-/- HeLa cells.
Interestingly, both CENP-E+/- and CENP-E-/- HeLa cells showed lagging chromosomes and chromosome breaks near the midbody during telophase. After CENP-E deletion, the structure of the midbody was altered, and the total fluorescence intensity of Aurora B at the midbody significantly increased. Additionally, the researchers found that CENP-E deletion led to an increase in micronuclei formation in both CENP-E+/- and CENP-E-/- HeLa cells.
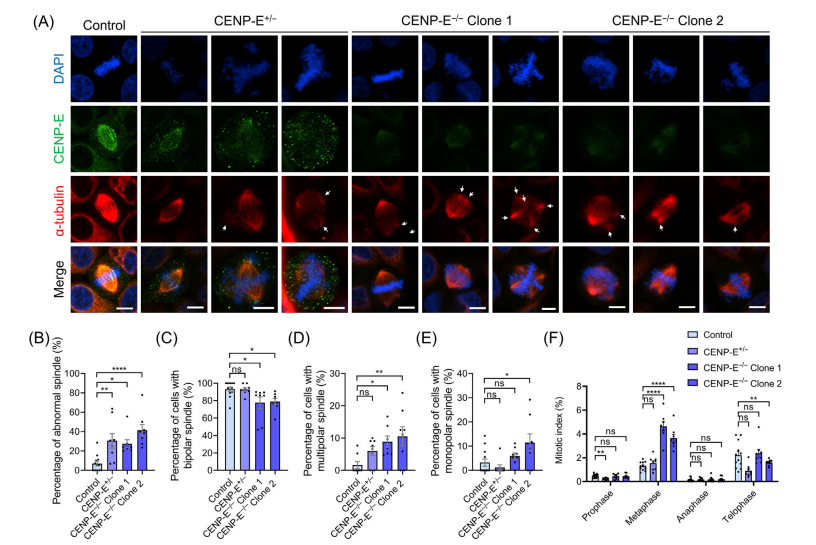
Fig.4 CENP-E deletion resulted in the formation of multipolar spindles and the disorganization of spindle poles.
In summary, as a plus-end-directed motor protein, CENP-E is essential for chromosome biorientation, congression, and alignment during cell division, ensuring error-free chromosome segregation and genome stability. With CRISPR-Cas9 knockout of CENP-E, the researchers observed chromosome misalignment, activation of the spindle assembly checkpoint, and subsequent cell cycle arrest, contributing to aneuploidy and genomic instability.
The study highlighted the co-dependence of CENP-E with key kinetochore proteins, such as BubR1 and KIF2C, and explored the regulatory roles of Aurora kinases in promoting chromosome congression. The findings underscored the critical role of CENP-E in maintaining spindle assembly checkpoint integrity, with its inhibition or deletion contributing to genomic instability, which can either suppress tumors or promote tumorigenesis. However, further research is needed to explore the molecular mechanisms of CENP-E's interactions with kinetochore proteins, as well as the survival pathways of CENP-E knockout cell lines.
Additionally, the inhibition of CENP-E through GSK923295 led to its disassociation from kinetochores and accumulation around spindle poles, highlighting its role in spindle pole maintenance. Both CENP-E inhibition and knockout resulted in an increase in multipolar spindles and centrosome dispersion, suggesting its involvement in bipolar spindle formation.
Collectively, with the technology of CRISPR-Cas9 gene editing, the researchers were able to underscore the essential role of CENP-E in spindle microtubule organization and the maintenance of spindle poles, contributing to proper chromosome alignment and congression, which provided a novel insight into cell division.
See how you can utilize CRISPR library screening technology in your research for target genes and pathways. Check out EDITGENE’s complete solution for CRISPR library screening, CRISPR KO/CRISPRa/CRISPRi, and the most extensive collection of library plasmids/viruses, which is available within weeks.
Recent Blogs:
1.[Weekly News] CRISPR screening reveals new mechanism: IL-4 contributes to CD8+ CART cell failure
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






