[Star of the Month] Yeo Lab RNA-Binding Protein CRISPR, Whole Genome Knockout, and Suppression Libraries

CRISPR Library Screening is a high-throughput gene screening approach based on CRISPR/Cas9 technology. It involves constructing a library containing tens of thousands of sgRNAs, which are then cloned into lentiviral vectors and used to infect target cells at a low MOI. This method ensures that each cell is infected with only one sgRNA, allowing for functional screening of genes corresponding to different sgRNAs.
In this regard, I would like to recommend three best-selling libraries from EDITGENE, along with interpretations of relevant research literature, with the hope that this information will contribute to advancing your research endeavors.
I. CRISPR KO Screening Identifies RNA-Binding Protein Eef1a1 as a Key Regulator of Myogenesis
Original Article Link: https://doi.org/10.3390/ijms25094816
Skeletal muscle myogenesis is a complex gene regulatory process, with the roles of transcription factors and non-coding RNAs well-studied. However, the role of RNA-binding proteins (RBPs) in muscle generation remains unclear. The CRISPR-Cas9 gene editing system has proven highly effective in conducting high-throughput loss-of-function screens, which are essential for exploring genetic factors associated with drug and toxin resistance, cancer metastasis, immune response, and more.
In this study, researchers utilized the CRISPR-Cas9 system to construct an RBP-KO (RNA-Binding Protein Knockout) library targeting 1,542 genes with 11,138 sgRNAs. Through screening, they identified Eef1a1 as a novel regulator critical for the proliferation of C2C12 myoblasts.
Subsequently, they successfully established C2C12 cell models with Eef1a1 knockout and knockdown using CRISPR KO and CRISPRi techniques, allowing for functional studies on Eef1a1. The results revealed that miR-133a-3p targets Eef1a1. Further investigation into the interaction between Eef1a1 and miR-133a-3p demonstrated that overexpression of miR-133a-3p could rescue the effects of Eef1a1 on C2C12 myoblasts.
In summary, the Yeo Lab RNA-Binding Protein Library screening revealed the critical role of Eef1a1 in myogenesis and elucidated the molecular mechanism by which it regulates muscle generation through miR-133a-3p.
This not only enhances our understanding of the molecular mechanisms of muscle generation but also provides new insights and potential targets for future clinical treatments and biotechnological applications.
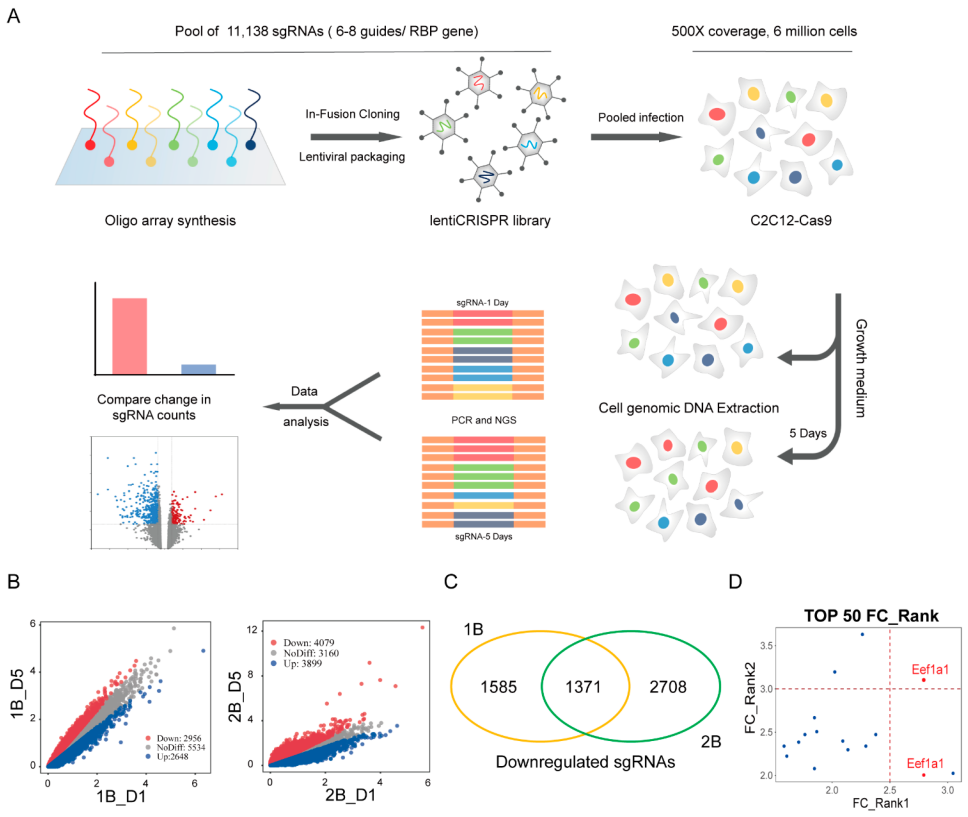
Figure 1: CRISPR Screening Shows Eef1a1 Involvement in Cell Proliferation
II. CRISPR KO Screening Identifies Therapeutic Targets for Translocation Renal Cell Carcinoma (tRCC)
Original Article Link: https://doi.org/10.1101/2024.08.09.607311
Translocation renal cell carcinoma (tRCC) is an aggressive subtype of renal cancer driven by TFE3 gene fusions. Unlike most other renal cell carcinomas (RCC), tRCC exhibits a metabolic profile characterized by oxidative phosphorylation (OXPHOS) rather than the typical aerobic glycolysis (Warburg effect).
Due to the unique biological characteristics of tRCC, conventional therapeutic drugs used for other RCC histological types have limited efficacy in tRCC, necessitating the development of new therapeutic strategies targeting the specific biology of tRCC.
In this study, researchers conducted a genome-scale CRISPR KO screening and identified selective vulnerabilities related to the metabolic state of tRCC cells, particularly the EGLN1 gene. EGLN1 encodes an enzyme that hydroxylates HIF-1α, marking it for degradation. Inhibiting EGLN1 stabilizes HIF-1α and promotes a metabolic reprogramming from OXPHOS to glycolysis, impacting the growth of tRCC cells.
Subsequently, the researchers performed functional validation experiments, drug sensitivity tests, and metabolic state analyses. They found that the TFE3 fusion protein not only promotes OXPHOS but also activates genes associated with glutathione and NADPH biosynthesis, creating a highly reduced cellular environment that may render tRCC sensitive to reductive stress.
In summary, through CRISPR screening using the Human Whole Genome Knockout Library , the researchers revealed the unique biological characteristics of tRCC and identified potential therapeutic targets, providing a scientific basis for the development of new treatment strategies.
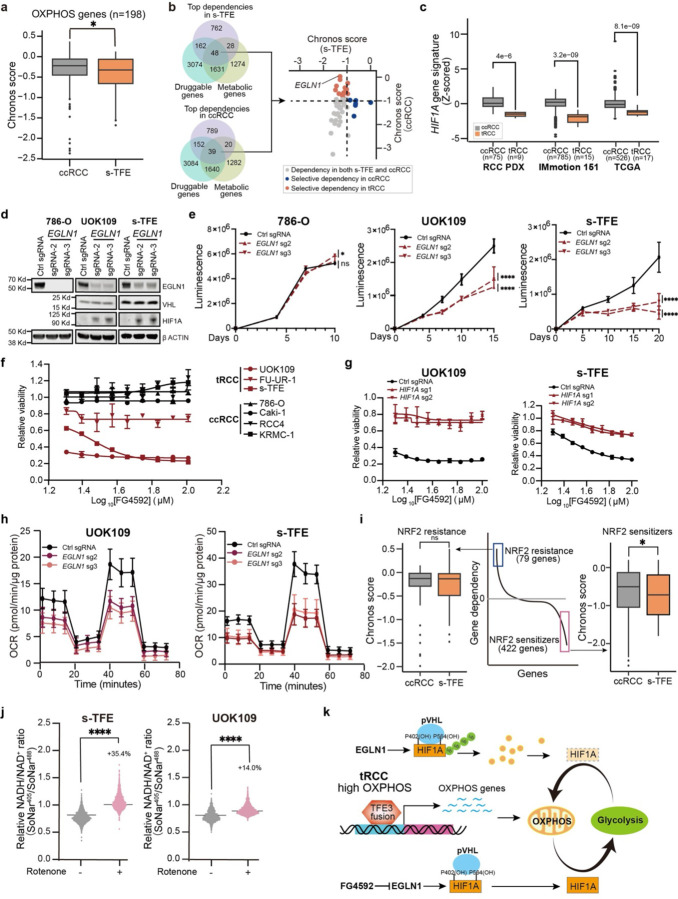
Figure 2: Selective EGLN1 Dependency in tRCC
III. CRISPRi Screening Reveals IRF9 as a Novel Biomarker for Sensitivity to DNA-Damaging Agents
Original Article Link: https://doi.org/10.3390/genes15070959
Poly(ADP-ribose) polymerase (PARP) inhibitors are a class of targeted therapeutics that interfere with DNA repair mechanisms to accumulate DNA damage and have been approved for the treatment of various cancers with BRCA1/2 mutations. However, resistance to PARP inhibitors has been observed in many cases over time. To overcome this resistance and identify biomarkers beyond BRCA1/2, researchers have been actively searching for new approaches.
Recent advancements in RNAi and CRISPR gene-editing technologies have identified numerous biomarkers related to the efficacy of PARP inhibitors and suggested new therapeutic targets for synthetic lethal strategies involving PARP inhibitors.
In this study, the research team used CRISPRi screening technology to identify genes that regulate sensitivity to PARP inhibitors in breast cancer cell lines. The experiments involved cell culture, gene editing, colony formation assays, qRT-PCR, and Western blotting.
The results demonstrated that the expression of the IRF9 gene is crucial for breast cancer cell sensitivity to PARP inhibitors; its loss leads to resistance to these inhibitors. Moreover, the upregulation of IRF9 expression after chemotherapy was associated with sensitivity to platinum-based chemotherapeutic agents. The upregulation of IRF9 may serve as a novel biomarker for predicting breast cancer patient sensitivity to both PARP inhibitors and platinum-based chemotherapy.
In summary, the researchers employed a Human Whole Genome Suppression Library to perform CRISPRi screening, ultimately revealing the potential importance of IRF9 in breast cancer treatment. This finding provides new perspectives and potential biomarkers for future cancer therapy strategies.
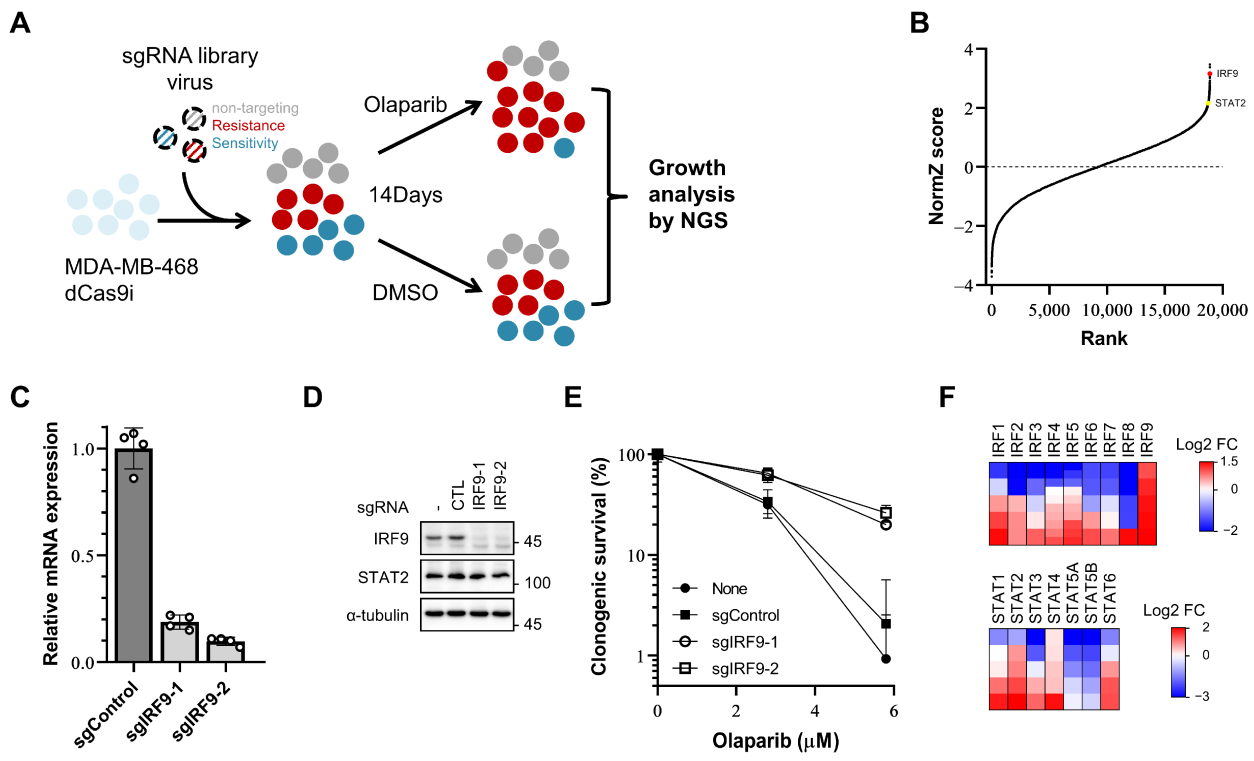
Figure 3: CRISPR/dCas9 interference screening identifies IRF9 and other IRF/STAT family members as resistance-inducing genes in TNBC cells.
EDITGENE, specializing in gene editing for over a decade, assembled the most popular types of CRISPR sgRNA libraries in the research community, with a coverage rate of ≥99% and uniformity <10. In-stock items are available within a week!
Recent Blogs:
1.[Weekly News] A Novel type VII CRISPR-Cas system: Discover the Potential of CRISPR Gene Editing
3.[Literature Review] SATCAS: A CRISPR/Cas13a-based isothermal one-pot RNA detection platform
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






