[Literature Review] CRISPR/Cas9 Cellular Reporter System to Evaluate Fetal Hemoglobin Induction and Therapeutic Screening

Reactivation of fetal hemoglobin (HbF) expression has long been a focal point in drug discovery efforts for treating β-globin disorders. With the advent of CRISPR gene therapy, reactivating HbF expression has emerged as a promising therapeutic approach.
To test potential HbF inducers, there is an urgent need for a sensitive, specific, and high-throughput screening system capable of quickly identifying potential HbF inducers among a large number of compounds. Driven by this need, many researchers aim to create a novel cellular reporter system to report endogenous HbF expression.
Recently, a group of researchers from the Netherlands, Germany, and Japan successfully developed such a cellular reporter system, with their findings published in the journal HemaSphere (IF: 12.1) under the title "A cellular reporter system to evaluate endogenous fetal hemoglobin induction and screen for therapeutic compounds."

Original Article Link:https://doi.org/10.1002/hem3.139
Globin disorders are a group of monogenic diseases, including sickle cell disease (SCD) and β-thalassemia, whose symptoms appear after birth when fetal hemoglobin (α2, β2) is replaced by adult hemoglobin (α2, β2). The switch from γ to β hemoglobin was observed in 1948 and was associated with the onset of symptoms. Since then, reactivating HbF expression has been considered a potential effective treatment for these patients.
Hydroxyurea is the only compound approved as a fetal hemoglobin inducer, but its efficacy varies widely among patients, with some patients showing no or limited response, highlighting the urgent need for new drug development.
Previous studies lacked a cell line capable of reporting endogenous HbF expression, which limited the evaluation and screening of new inducers. In this study, researchers utilized CRISPR/Cas9 technology to insert a bioluminescent tag into the adult erythroid progenitor cell line HUDEP2, creating the first endogenous HbF reporter cell line.
Finally, the researchers used this novel reporter cell line to evaluate the effects of genetic and pharmacological strategies on inducing fetal hemoglobin expression and to identify potential HbF inducers through high-throughput drug screening.
I. Sequence Analysis of HBG1-Specific CRISPR-Cas9 gRNA Design
The fetal γ-globin chain is encoded by two nearly identical and closely spaced genes, HBG1 and HBG2. Due to the high sequence homology between these two genes, Cas9-induced double-strand breaks could potentially occur at both loci, leading to large deletions or inversions. Therefore, for CRISPR-Cas9-mediated endogenous tagging of the fetal γ-globin chain, it is crucial to design gene-specific gRNAs that target HBG1 without affecting other β-like globin genes at the HBB locus.
To design a CRISPR-Cas9 gRNA specifically targeting the HBG1 gene (which encodes the Aγ-globin chain), the researchers first analyzed the relevant sequences. They compared the 3'-UTR regions of HBG1 and HBG2 (which encodes the Gγ-globin chain) in the human reference genome GRCh38, as well as gene-specific PCR amplicon sequencing in HUDEP2 cells.
The analysis revealed a unique nucleotide at position +17 and an additional adenine at position +55 in the 3'-UTR of HBG1. In HUDEP2 cells, other gene-specific nucleotides were also identified in the 3'-UTR. Additionally, the extra adenine at position +55 in HUDEP2 cells was found to be allele-specific rather than gene-specific. Using this sequence information, the researchers designed two gRNAs specifically targeting the HBG1 3'-UTR.
By cloning these gRNAs into expression plasmids and transfecting them into HEK293T cells, the researchers evaluated their DNA cleavage efficiency. Using the TIDE algorithm to analyze Sanger sequencing traces, they found that gRNA #2 exhibited the highest efficiency.
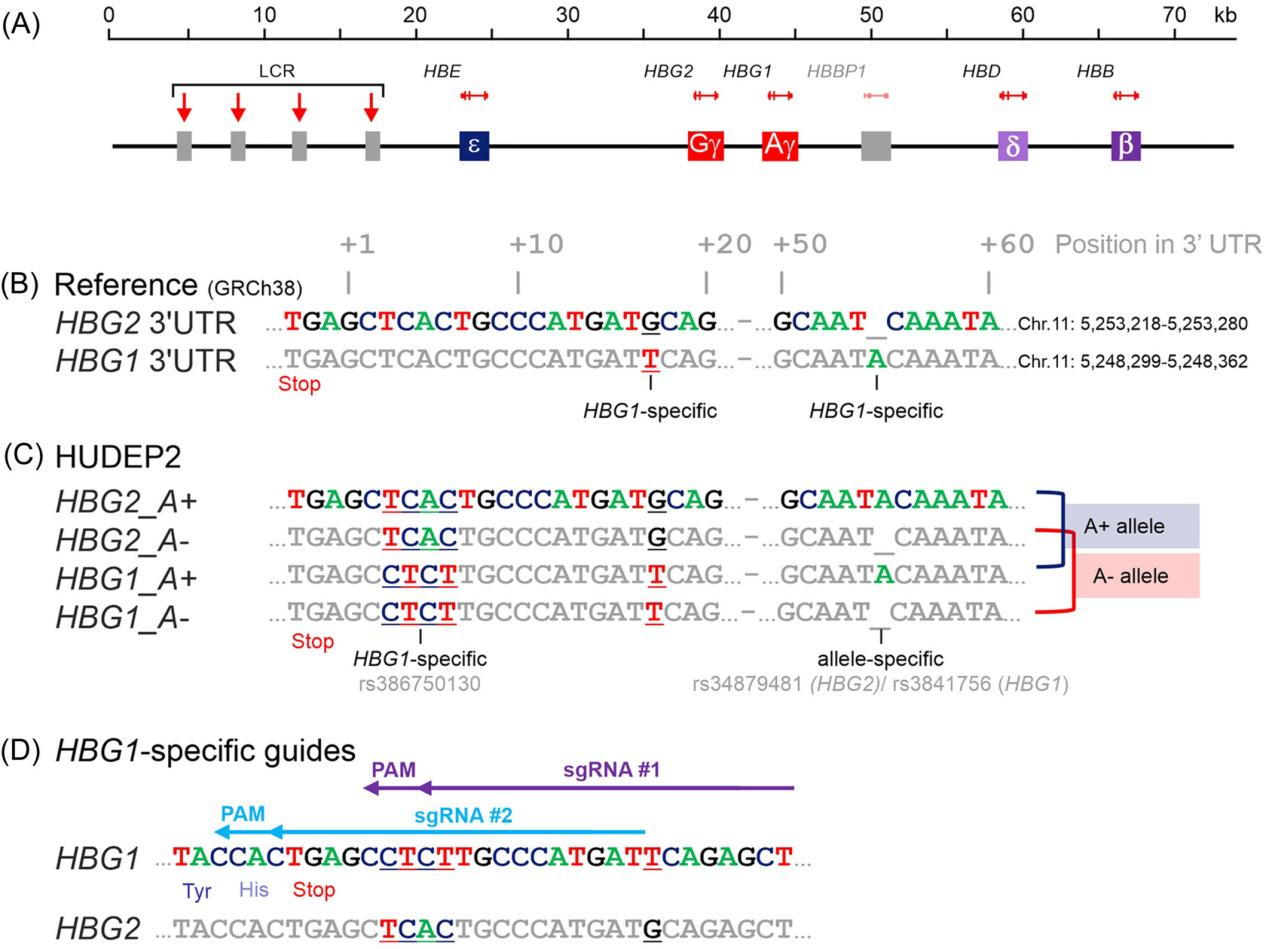
Figure 1. Design of gRNAs Specifically Targeting the HBG1 3'-UTR in HUDEP2 Cells
II. Tagging the Endogenous HBG1 Gene in HUDEP Cells
Before specifically tagging the endogenous HBG1 gene in the HUDEP cell line, researchers first tested the feasibility of CRISPR-Cas9-mediated HBG1 gene tagging in fetal-like HUDEP1 cells. They co-transfected a double-stranded DNA template containing the eGFP reporter gene with the Cas9_gRNA #2 plasmid, attempting to edit the HBG1 gene. Flow cytometry analysis revealed a low knock-in efficiency (GFP-positive cells were less than 1%). However, correctly edited cells could be enriched by fluorescence-activated cell sorting (FACS). DNA sequence analysis of the FACS-sorted cells confirmed that the eGFP tag was correctly inserted into the endogenous HBG1 gene.
To achieve a more efficient knock-in strategy, researchers used a single-stranded DNA template and an improved Cas9-gRNA delivery method (using RNP instead of plasmids) to introduce the HiBiT tag at the C-terminus of the Aγ-globin in HUDEP2 cells. With these optimizations, the knock-in efficiency increased to over 20%. Western blot analysis showed that in clone #10, both HBG1 genes contained the HiBiT tag, and the Aγ-HiBiT chain was detected as a slower-migrating band compared to the untagged Gγ chain. HiBiT luminescence signal detection of the western blot confirmed that only the slower-migrating Aγ-HiBiT band was detected, verifying the specificity of the HiBiT tag.
By repeating this process, the researchers generated a reporter cell line with the HiBiT tag in non-HbF-expressing HUDEP2 cells, which was validated through molecular characterization and high-throughput screening.
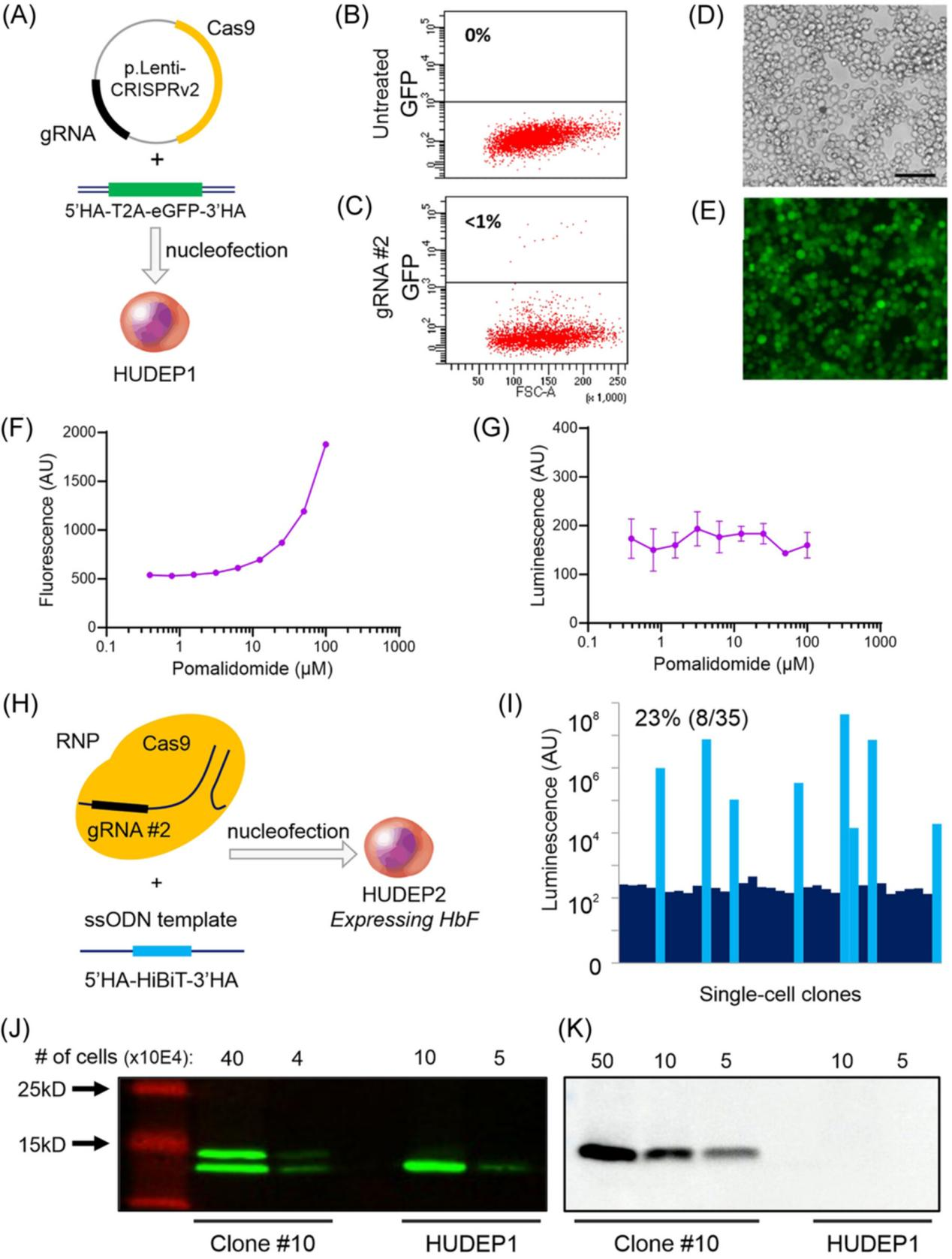
Figure 2. C-terminal Tagging of the HBG1 Gene
III. Aγ-HiBiT Reporter Cell Line Exhibits Similar Proliferation and Differentiation Characteristics as HUDEP2 Cells
To confirm whether the newly created Aγ-HiBiT reporter cell line exhibits biological behaviors similar to the original HUDEP2 cell line, particularly in the key processes of cell proliferation and differentiation, researchers conducted a series of validations. Sequence analysis confirmed that the Aγ-HiBiT reporter cell line had a HiBiT tag inserted at the A− allele of the HBG1 gene, while an insertion mutation preserving the TGA stop codon was observed on the A+ allele.
SNP array analysis showed that, in addition to the known trisomy of chromosomes 6, 8, 18, 19, and 21 in HUDEP2 cells, the reporter cell line had an additional copy of chromosome 7. ATAC-seq analysis revealed that the open chromatin pattern at the HBB locus in the reporter cell line was similar to that in HUDEP2 cells, and there was no ATAC signal in the HBE, HBG1, and HBG2 gene regions, indicating that these genes remained effectively silenced.
RNA sequencing results showed that transcriptional activity in the reporter cell line was limited to the adult HBD and HBB genes, similar to the expression pattern observed in HUDEP2 cells. Additionally, they compared the expression levels of 52 known HbF regulators in the reporter cell line and HUDEP2 cells and analyzed the phenotypic changes during the differentiation process. These experiments collectively confirmed that the Aγ-HiBiT reporter cell line exhibits a high degree of similarity to HUDEP2 cells at both the molecular and cellular levels.
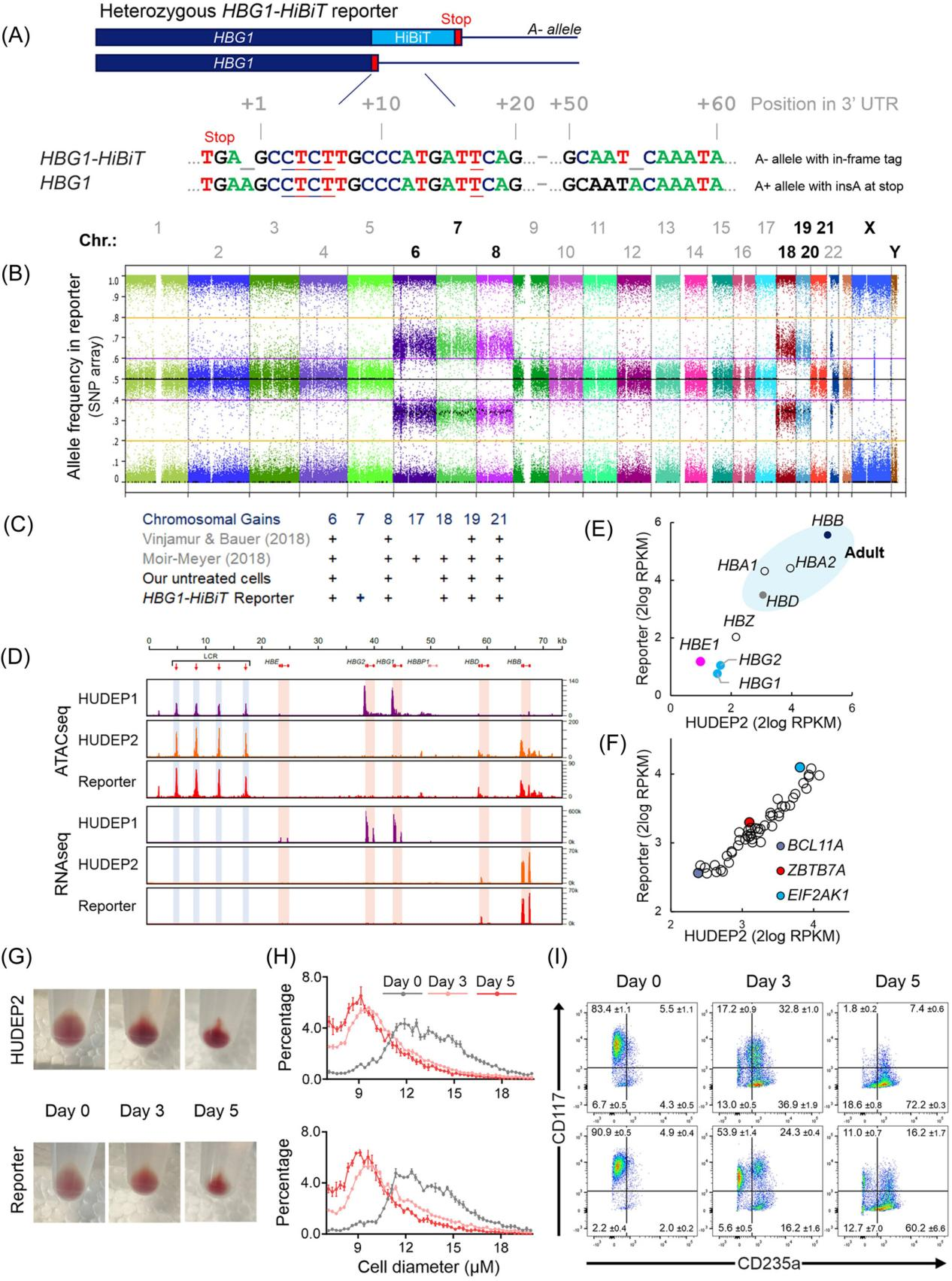
Figure 3. Characterization of the Aγ-HiBiT Reporter Cell Line
IV. Application of the Aγ-HiBiT Reporter Cell Line to Evaluate Genetic HbF-Induction Strategies
After creating the Aγ-HiBiT reporter cell line, to better understand and manipulate HbF expression and develop new therapeutic approaches, researchers further explored how to use the Aγ-HiBiT reporter cell line to evaluate genetic HbF (fetal hemoglobin) induction strategies.
The researchers first confirmed whether the Aγ-HiBiT reporter cell line could survive gene editing experiments. They used Cas9-guided gRNAs targeting the promoter regions of the HBG1 and HBG2 genes to introduce a known 13 bp deletion associated with HPFH (Hereditary Persistence of Fetal Hemoglobin) into the reporter cells via the CRISPR-Cas9 system.
In both the reporter cells and control HUDEP2 cells, an increase in γ-globin protein levels was observed after genome editing. Western blot analysis showed that in the reporter cells, both HiBiT-tagged and untagged γ-globin chains could be detected after gene editing. HPLC (high-performance liquid chromatography) analysis revealed that the proportion of HbF in the reporter cell population increased from approximately 2.3% to 23.1% after gene editing.
In addition to editing the HBG1/2 genes, the study also explored another genetic strategy: disrupting the erythroid-specific enhancer of BCL11A, a key repressor of HbF. The HiBiT Lytic Assay showed a significant increase in HiBiT signal in the reporter cells following both genetic approaches—HBG promoter editing and BCL11A enhancer disruption.
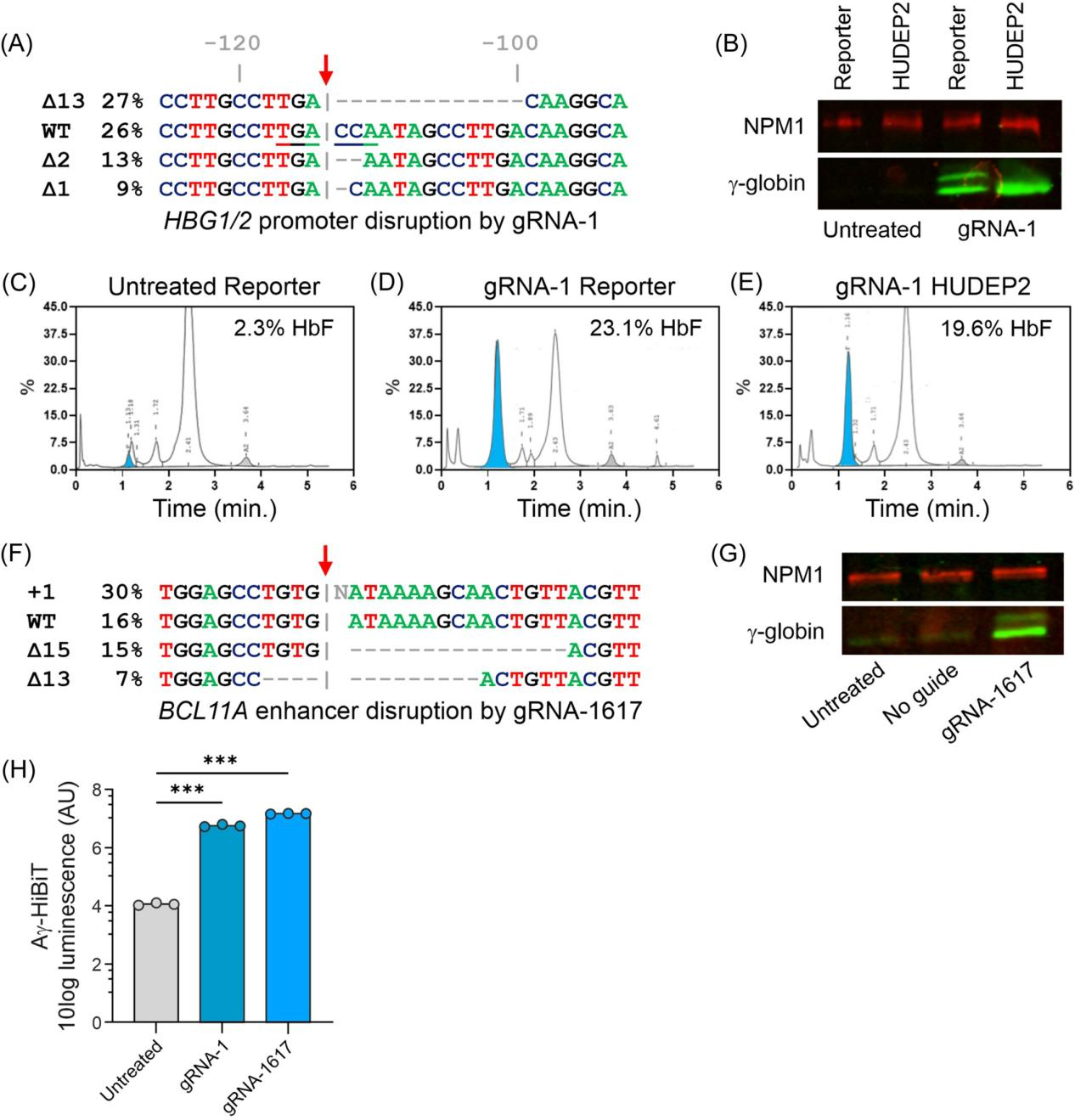
Figure 4. Using the Aγ-HiBiT Reporter Cells to Evaluate Genetic Induction of Fetal Hemoglobin (HbF)
V. Pomalidomide as a Positive Control Compound for HbF Induction
To discover compounds capable of inducing γ-globin expression, the researchers selected pomalidomide as a positive control compound for HbF induction. They treated primary erythroid progenitor cells from healthy donors and sickle cell disease patients with pomalidomide to evaluate its effect on HbF expression.
First, the researchers assessed the expression levels of γ-globin mRNA and protein after pomalidomide treatment using quantitative reverse transcription polymerase chain reaction (RT-qPCR) and western blot analysis. The results showed that pomalidomide treatment significantly increased HbF levels in primary erythroid progenitor cells, both under proliferative and differentiative conditions.
Moreover, when conducting compound screening, the impact of the compound on cell viability is an essential factor to consider. After determining that pomalidomide did not exhibit significant toxicity within a certain concentration range, the researchers tested the effect of various concentrations of pomalidomide on the Aγ-HiBiT signal. They observed a dose-dependent increase and estimated the EC50 value.
To verify whether pomalidomide specifically activates HbF, the researchers also tested its effect under differentiative conditions and compared it with cytotoxic compounds to ensure that HbF induction was not due to a cellular stress response.
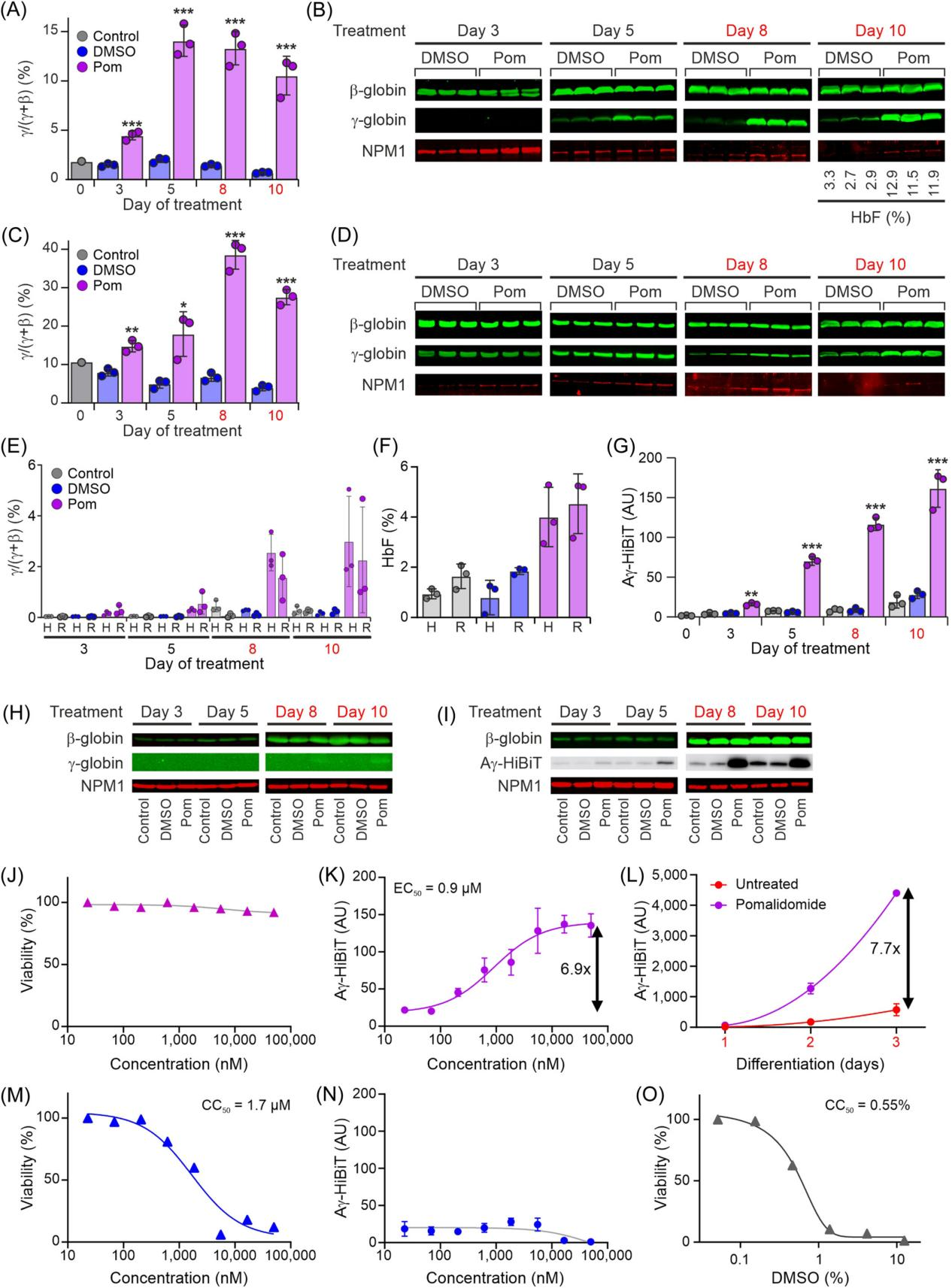
Figure 5. Using Pomalidomide as a Positive Control for Fetal Hemoglobin (HbF) Induction
VI. Compatibility of the Reporter Cell Line with High-Throughput Screening (HTS)
To verify whether the Aγ-HiBiT reporter cell line is suitable for high-throughput screening (HTS), the researchers conducted compatibility tests of the Aγ-HiBiT reporter cell line with HTS. After determining the optimal culture conditions for the Aγ-HiBiT reporter cell line suitable for HTS, the researchers tested different cell seeding densities in 384-well plates and found that 10,000 to 15,000 cells per well could proliferate linearly within three days.
Next, the researchers treated the cells with various concentrations of pomalidomide to test the dose-dependent response of the Aγ-HiBiT signal. To address issues that arose during automated readout, they optimized experimental procedures, including reducing the volume of lysis buffer and adjusting the addition speed.
By screening a library of 5,632 known compounds, the researchers validated the applicability of the Aγ-HiBiT reporter cell line in HTS. The screening successfully identified the known HbF inducer decitabine, as well as other nucleotide analogs, demonstrating the cell line's effectiveness in identifying HbF inducers.
Through DMSO solvent toxicity studies, the researchers determined the sensitivity of the Aγ-HiBiT reporter cell line to DMSO and recommended using low concentrations of DMSO in HTS to avoid affecting cell viability.
These experimental results ultimately confirmed that the Aγ-HiBiT reporter cell line is compatible with HTS and can serve as a powerful tool for the unbiased identification of new HbF-inducing compounds.
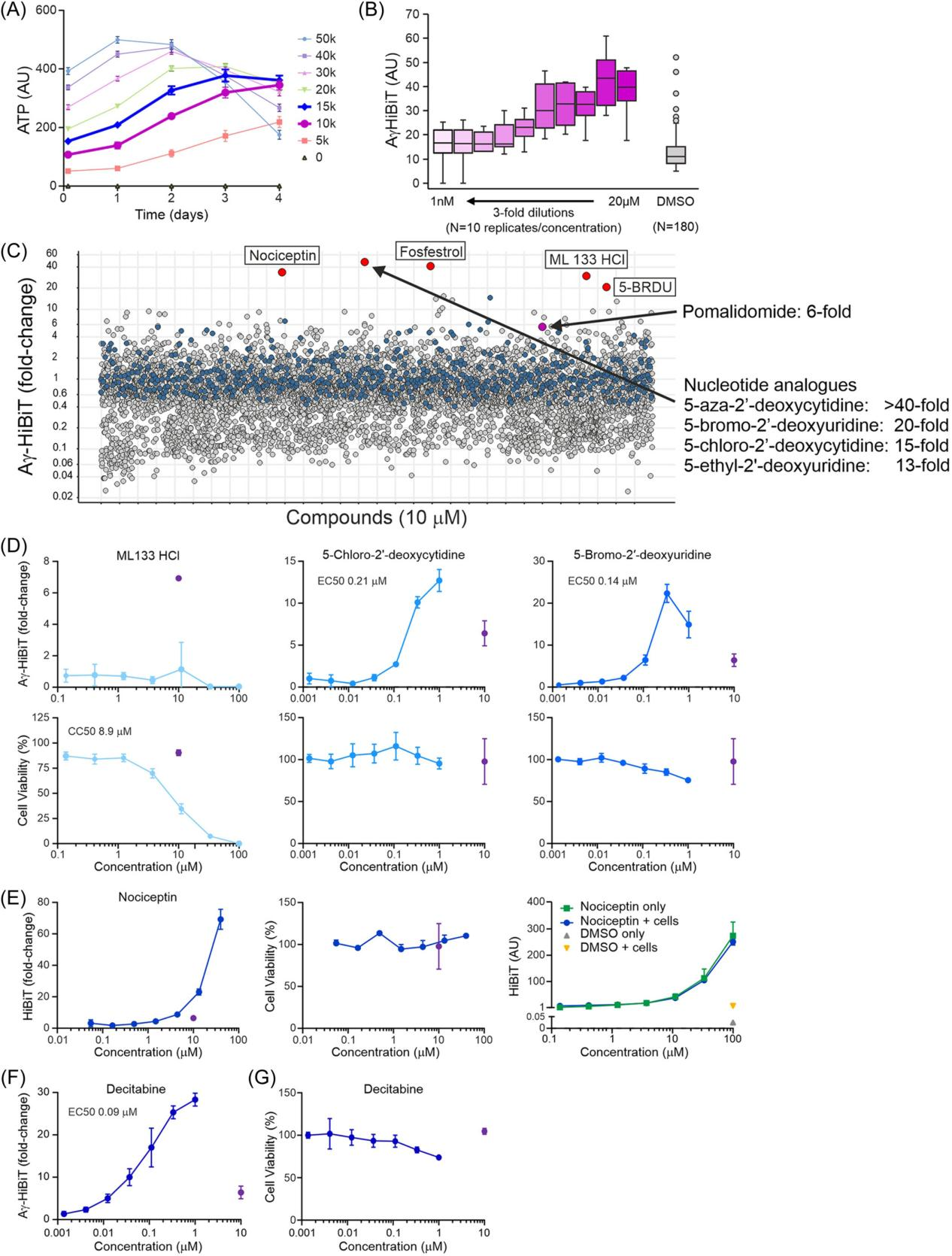
Figure 6. Aγ-HiBiT Reporter Cell Line Suitable for High-Throughput Screening (HTS) Applications Aimed at Detecting Hemoglobin (HbF) Induction
In summary, this study reports the development of an innovative Aγ-HiBiT reporter cell line, which uses CRISPR-Cas9 gene knock-in technology to specifically tag the fetal hemoglobin (HbF) gene in adult erythroid progenitor HUDEP2 cells.
This system enables highly sensitive and specific monitoring of endogenous HbF expression and is suitable for high-throughput drug screening to identify new HbF inducers. This work provides a new tool for the therapeutic research of β-globin disorders, contributing to the advancement of new treatment approaches.
EDITGENE has innovatively developed an efficient gene knock-in technology, offering customized gene knock-in services using an upgraded CRISPR/Cas9 system. With over a decade of experience in gene editing, EDITGENE has refined the optimal gRNA and homology arm design strategies, ensuring that their gene knock-in services deliver higher positive rates and broader applicability across various gene loci!
Recent Blogs:
1.[Literature Review] SATCAS: A CRISPR/Cas13a-based isothermal one-pot RNA detection platform
3.[Literature Review] CRISPR KO Screening Reveals Key Genes Responsible for Chemotherapy Resistance
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






