[Literature Review] CRISPR-Cas12a Exhibits Metal-Dependent Specificity Switching

Cas12a is an effector protein of the type V-A CRISPR-Cas immune system, and its specificity, due to potential off-target effects, has been a widely studied subject in in vitro and genome editing experiments.
Some previous reports have indicated that the specificity of Cas12a may change under physiological conditions. In particular, the concentration of free magnesium ions in cells is typically lower than the concentration used in in vitro studies. To this end, researchers sought to further investigate the differences in Cas12a specificity under these conditions.
Recently, researchers from Iowa State University in the United States published a paper titled "CRISPR-Cas12a Exhibits Metal-Dependent Specificity Switching" in Nucleic Acids Research. This study highlight the results of Cas12a exhibiting metal-dependent specificity switching.
Original Article Link: https://doi.org/10.1093/nar/gkae613
The specificity of Cas12a is primarily determined by two factors: the complementarity between crRNA and the DNA target, and the protospacer adjacent motif (PAM) located adjacent to the DNA target.
Although the specificity of Cas12a has been widely studied, many in vitro investigations have been conducted under high magnesium ion concentrations, which may not reflect the actual free Mg²⁺ concentrations present within cells.
To address this issue, researchers conducted analyses of the specificity of Cas12a homologs across a range of Mg²⁺ concentrations. The results demonstrated that Cas12a alters its specificity based on the concentration of metal ions. Specifically, at low Mg²⁺ concentrations, Cas12a exhibited increased tolerance for mismatches in the seed region while displaying decreased tolerance for mismatches at the PAM-distal end.
I. The Influence of Mg²⁺ Concentration on Off-Target Effects and Trans-Cleavage Activity of Cas12a
Previous studies have indicated that phages can evade CRISPR-mediated immune responses by generating mutations in the PAM and seed regions. However, researchers have observed that multiple mismatches in the PAM-distal region can also enable phages to preferentially escape from Cas12a.
Through head-to-head competition assays, they discovered that different types of phage mutants exhibited varying population distribution patterns based on the degree of crRNA matching.
Specifically, when using crRNA with complete complementarity, seed region mutants predominated; whereas with crRNA having PAM-distal mismatches, PAM-distal mutants began to dominate.
Researchers further observed that at low MgCl₂ concentrations (1 mM), the cleavage rate for seed region mutants was significantly faster compared to PAM-distal mutants, which is the opposite of what was observed at high MgCl₂ concentrations (10 mM).
To further analyze the specificity of Cas12a under different Mg²⁺ concentrations, researchers conducted in vitro plasmid library cleavage experiments. In these experiments, Cas12a-crRNA-protein complexes were used to cleave plasmid libraries containing mutant target sequences. Samples were collected at various time points and analyzed via agarose gel electrophoresis to assess cleavage results.
The experimental outcomes demonstrated that Cas12a's tolerance for seed region mutations varied with Mg²⁺ concentration, showing higher tolerance at low Mg²⁺ levels, while tolerance for PAM-distal region mutations decreased.
Furthermore, the non-specific DNA cleavage activity of Cas12a, activated by the target sequence, was more pronounced at high Mg²⁺ concentrations but diminished at low Mg²⁺ levels. These experimental results indicate that both the specificity and non-specific cleavage activity of Cas12a are significantly influenced by Mg²⁺ concentration.
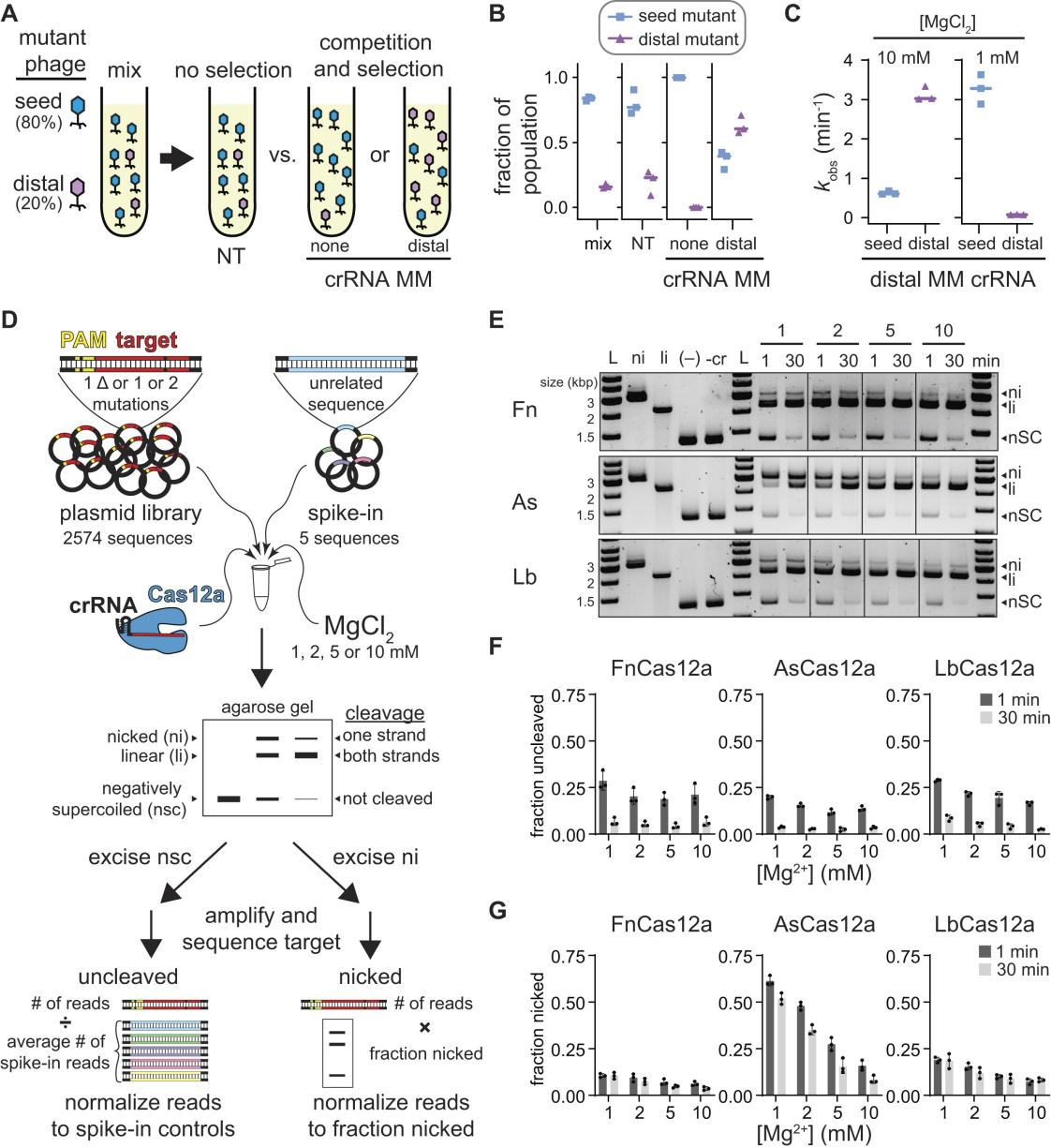
Figure 1: Analysis of Mg²⁺-Dependent Cas12a Specificity Using Plasmid Library Cleavage
II. Effects of Different Mg²⁺ Concentrations on PAM-Proximal and Distal Mutants
To understand how Mg²⁺ concentration affects Cas12a's recognition and cleavage of different types of mutants, researchers conducted further studies using analytical tools such as volcano plots and heatmaps.
Through volcano plot analysis, they compared Cas12a cleavage of uncleaved or nicked sequences under 1 mM and 10 mM MgCl₂ conditions. The volcano plots revealed that the first mutation in the PAM or seed region is more likely to remain uncleaved at high MgCl₂ concentrations, while the first or second mutation in the middle or PAM-distal region is more likely to remain uncleaved or form nicks at low MgCl₂ concentrations.
The heatmap analysis further illustrated the abundance of various mutant sequences in uncleaved and nicked fractions under different Mg²⁺ concentrations.
Additionally, researchers explored the mechanism of Cas12a nicking, finding that specific mutation combinations at high Mg²⁺ concentrations led to nicking, whereas at low Mg²⁺ concentrations, at least one mutation in the middle or PAM-distal region resulted in nicking.
Experiments on the metal ion dependence of Cas12a revealed that the second-strand cleavage step of Cas12a is significantly affected at low Mg²⁺ concentrations, especially for AsCas12a, indicating that Cas12a's cleavage activity is metal ion-dependent.
These findings demonstrate that changes in Mg²⁺ concentration have a significant impact on Cas12a specificity; at low Mg²⁺ concentrations, Cas12a is more tolerant of mutations proximal to the PAM, while its tolerance for distal mutations decreases.
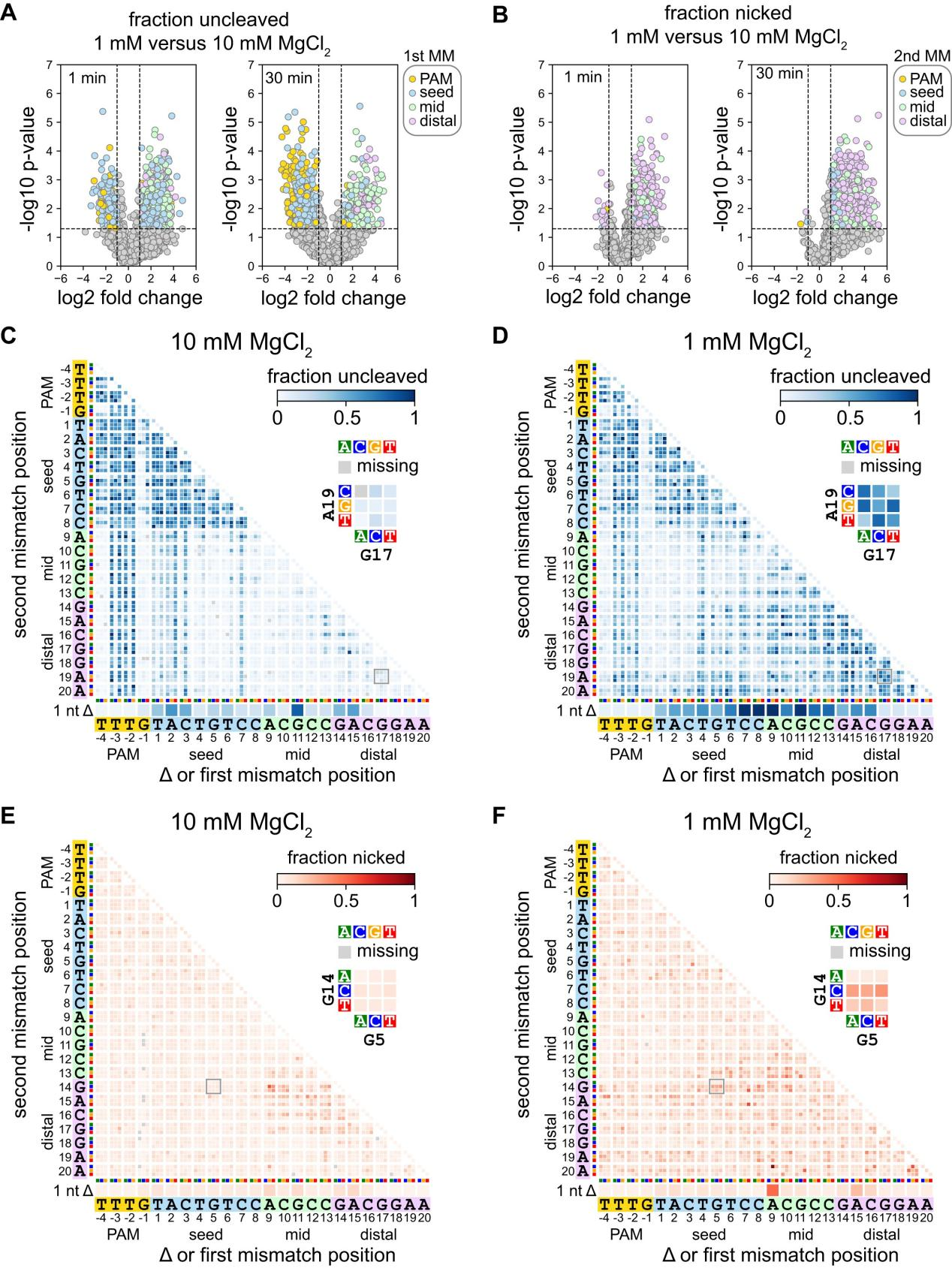
Figure 2: Specificity in the Seed and PAM-Distal Regions Changes at Low Mg²⁺ Concentrations
III. Visualizing Metal-Dependent Specificity Switch
To visually demonstrate the impact of Mg²⁺ on Cas12a specificity, researchers conducted experiments to observe and quantify changes in Cas12a specificity under different metal ion concentrations, presenting the results in graphical form.
They set two time points (1 minute and 30 minutes) to observe Cas12a's cleavage efficiency at different Mg²⁺ concentrations and used gel electrophoresis to separate uncleaved (supercoiled), single-strand nicked (nicked), and fully cleaved (linear) DNA.
By calculating the fractions of each mutant sequence that remained uncleaved or single-strand nicked at different Mg²⁺ concentrations, the researchers estimated the proportion of fully cleaved DNA.
Utilizing these data, they created volcano plots and heatmaps, visualizing the effect of Mg²⁺ concentration changes on Cas12a cleavage specificity. These charts revealed changes in the fractions of uncleaved or single-strand nicked sequences of specific mutant types at low Mg²⁺ concentrations.
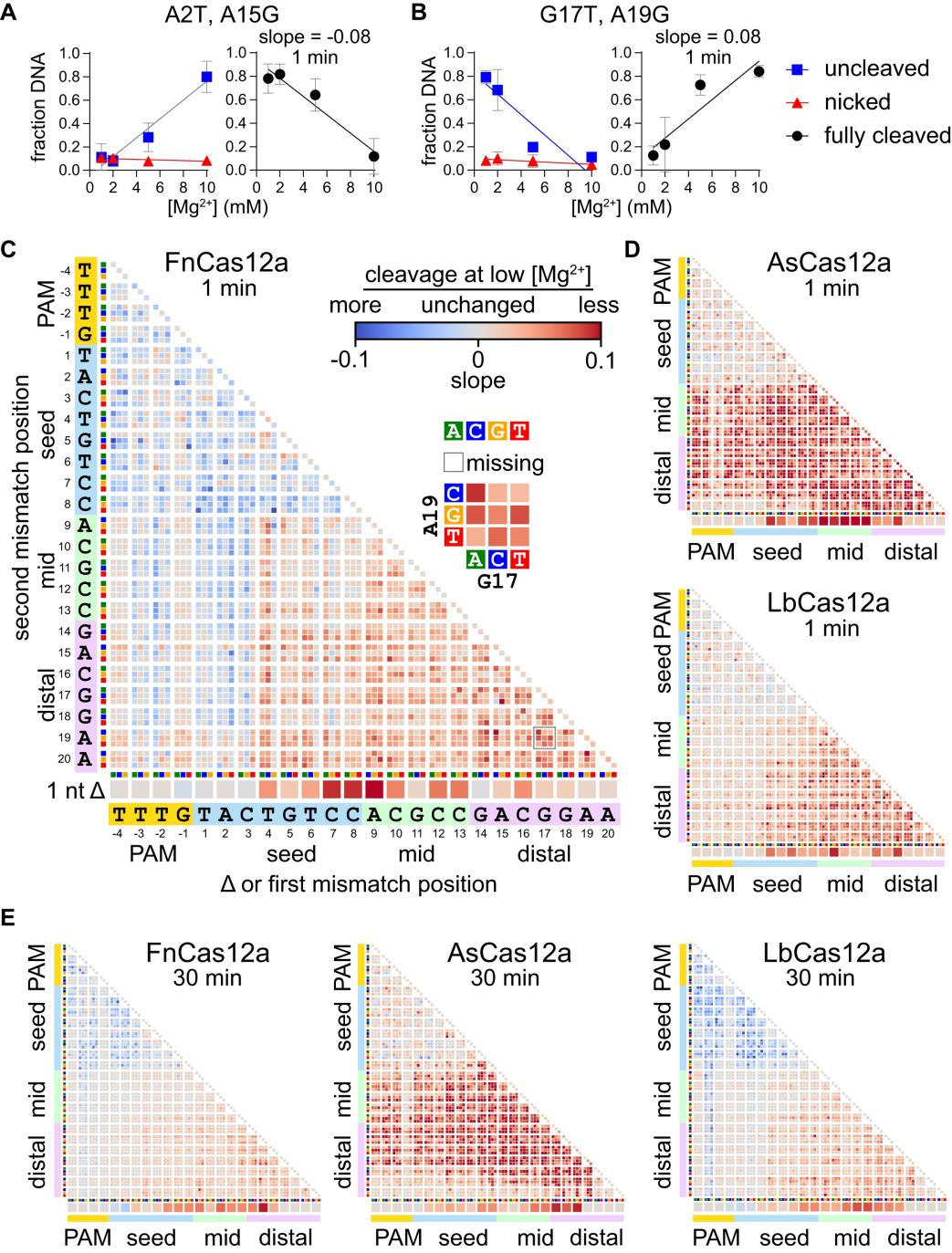
Figure 3: Metal-Dependent Specificity Switch in Three Cas12a Homologs
IV. Different Mechanisms of Mg²⁺-Dependent Specificity Switch
To investigate how Mg²⁺ affects Cas12a specificity differently depending on mutation location, researchers further studied FnCas12a, analyzing the cleavage mechanism by initiating the cleavage reaction in two ways: mixing the RNP with DNA in the presence of Mg²⁺, and pre-mixing the RNP and DNA without Mg²⁺ and then initiating cleavage by adding Mg²⁺.
The results showed that when the cleavage reaction was initiated by adding Mg²⁺, the first-strand cleavage of fully matched and seed-mutated targets was completed within 7 seconds. PAM-distal mutant targets also showed rapid first-strand cleavage at 10 mM Mg²⁺ but significantly reduced cleavage rates at 1 mM Mg²⁺.
Furthermore, researchers measured the rate constants of the cleavage reactions under different conditions to evaluate the rate-limiting steps. The results indicated that for seed-mutated targets, DNA binding was the rate-limiting step.
In contrast, for PAM-distal mutant targets at low Mg²⁺ concentrations, the cleavage defect might occur in the steps after DNA binding but before the cleavage of each strand, possibly due to inhibited necessary conformational changes.
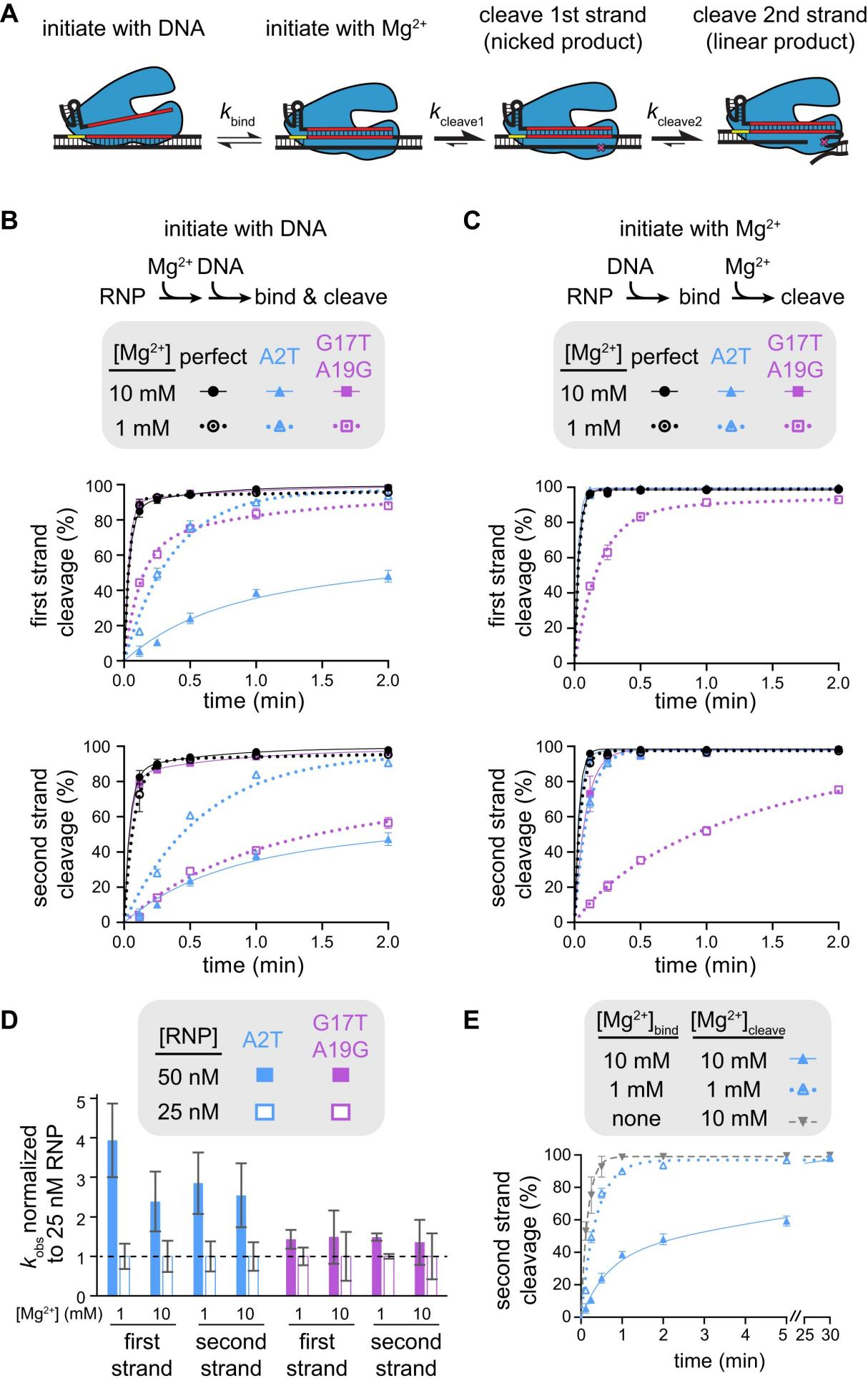
Figure 4: Different Mechanisms of Specificity Switch for Seed and PAM-Distal Mutants
V. Phage Escape Results Reveal the Consequences of Cas12a Homolog Specificity
Researchers discovered that different Cas12a homologs (such as AsCas12a, FnCas12a, and LbCas12a) exhibit varied specificity at low Mg²⁺ concentrations. However, how this specificity affects phage escape during bacteriophage challenges was not clear.
To investigate, the researchers conducted experiments by expressing different Cas12a homologs and crRNA in E. coli, followed by λ phage infection. From cultures treated with AsCas12a and FnCas12a, they found phage escape mutants.
The phage populations treated with AsCas12a displayed the highest mutation diversity, while those treated with Fn and LbCas12a mainly showed mutations at the most detrimental positions in the PAM and seed regions.
Furthermore, an analysis of the mutation locations in the phage populations revealed that phages treated with AsCas12a had widespread mutations in the PAM and seed regions, whereas those treated with Fn and LbCas12a predominantly had mutations at specific positions within the seed region.
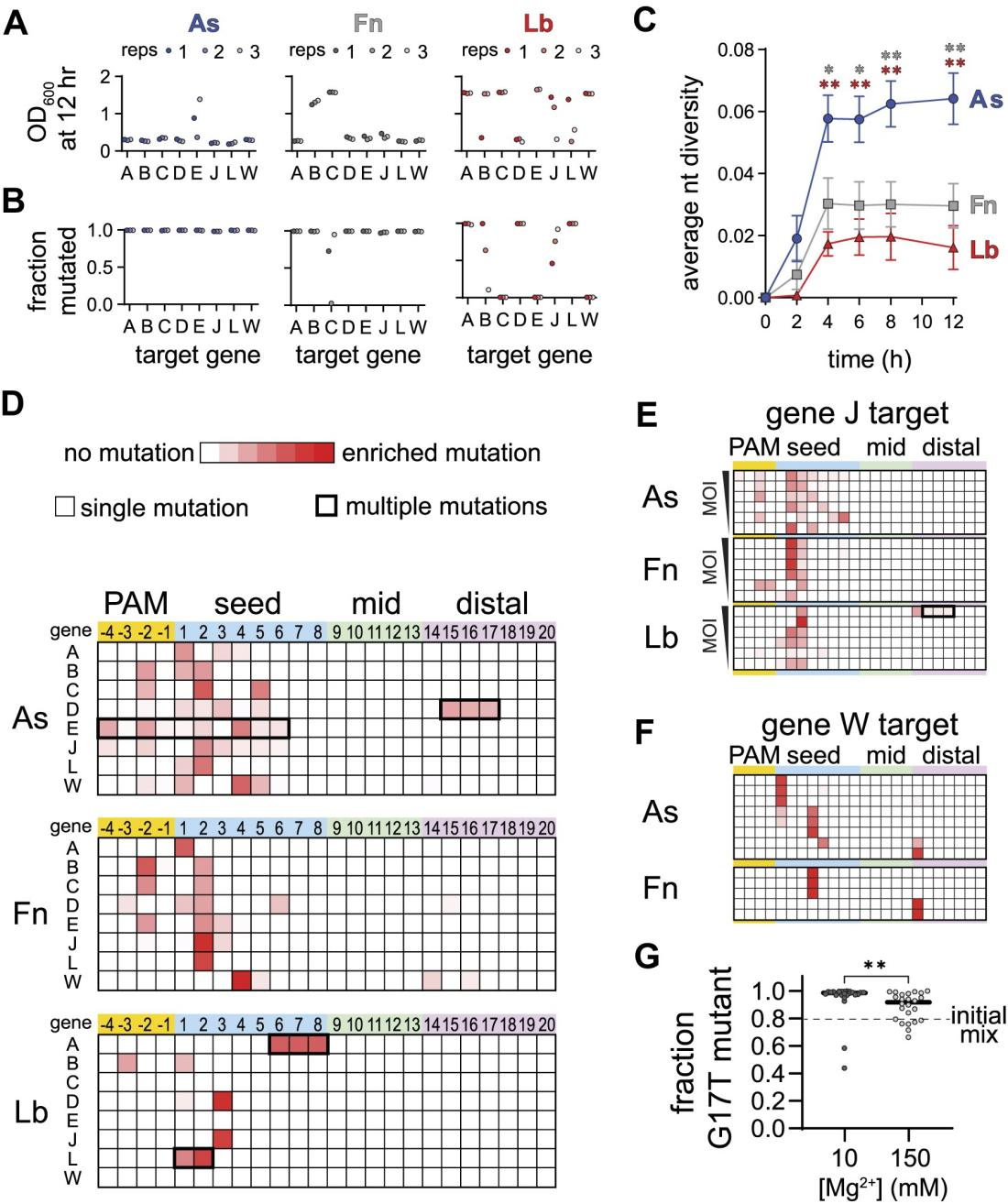
Figure 5: Different Escape Outcomes of Cas12a Homologs
VI. Metal-Dependent Specificity Can Alter Phage Escape Outcomes
After determining that Cas12a enzyme specificity is influenced by its homolog type, researchers further explored how metal ion concentration affects this specificity and phage escape outcomes.
They repeated the phage competition experiments in media supplemented with different Mg²⁺ concentrations. The results showed that under 10 mM Mg²⁺ conditions, PAM-distal mutant phages dominated in nearly all cultures, whereas their dominance significantly decreased under 150 mM Mg²⁺ conditions.
These findings indicate that changes in intracellular Mg²⁺ concentration can significantly influence phage escape outcomes. At low Mg²⁺ concentrations, Cas12a is more tolerant of seed mutations, while its tolerance for PAM-distal mutations decreases.
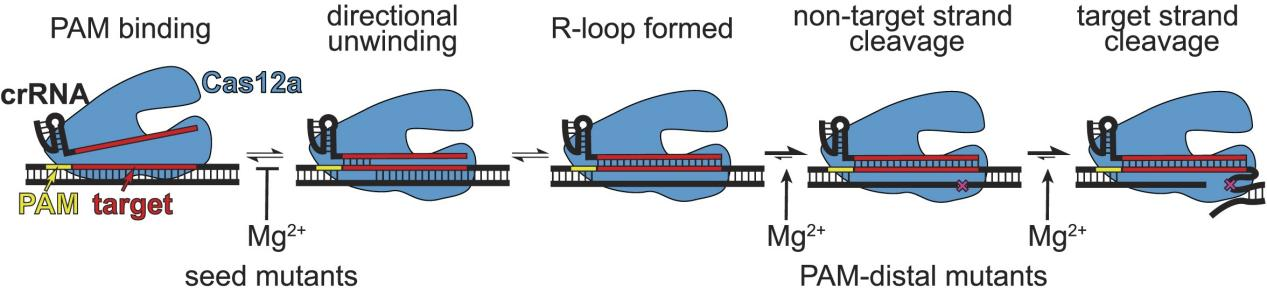
Figure 6: Model of Mg²⁺-Dependent Cas12a Steps and Their Differential Impact Based on Mutation Sites
In summary, researchers conducted a series of experiments to study the specificity changes of CRISPR-Cas12a enzymes under different Mg²⁺ ion concentrations and explored how these changes affect phage escape and genome editing.
The results indicate that Cas12a alters its specificity based on metal ion concentration, with lower Mg²⁺ concentrations reducing cleavage defects caused by seed mismatches while increasing defects caused by PAM-distal mismatches.
Significant differences in specificity were observed among different Cas12a homologs at varying Mg²⁺ concentrations, leading to distinct mutation types and diversity in the phage populations they processed. These differences altered the outcomes of phage escape.
This study is significant for understanding and optimizing the application of CRISPR-Cas systems in genome editing.
EDITGENE boasts its own independently developed protein purification platform. Our Cas enzymes have shown significantly superior activity compared to NEB and other market enzymes through active testing, achieving the highest standards in purity, activity, and sensitivity.
Be on the lookout for our isothermal amplification kit and CRISPR detection kits, efficient with minimal equipment requirements and short reaction times—Coming soon!
Recent blogs:
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






