[Literature Review] Multiplex Detection Strategy of Biosensors Based on CRISPR-Cas System
In recent years, the CRISPR-Cas system has been widely used in the field of nucleic acid detection due to its strong specificity, high sensitivity, and excellent cleavage activity. To obtain more valuable information within limited time and cost, many researchers have focused on achieving multiplex nucleic acid detection in a single CRISPR-Cas reaction. This need has become particularly urgent during the COVID-19 pandemic, as various mutations of SARS-CoV-2 have emerged, highlighting the necessity for CRISPR-Cas multiplex detection to support point-of-care testing (POCT).
Associate Professor Yu Songcheng's team from Zhengzhou University in China has recently published a review article titled "Multiplexed Detection Strategies for Biosensors Based on the CRISPR-Cas System" in the journal ACS Synthetic Biology. This review summarizes the principles and characteristics of current multiplex detection strategies based on the CRISPR-Cas system, analyzes potential challenges, and explores future developments.

Original Article Link: https://pubs.acs.org/doi/10.1021/acssynbio.4c00161?ref=PDF
CRISPR-Cas biosensors are characterized by short reaction times and signal amplification, but their non-specific cleavage activity makes it difficult to distinguish multiple targets in a single reaction. Despite this, many researchers have achieved multiplex target detection based on CRISPR-Cas through various strategies. Depending on the methods used, these multiplex detection strategies include microfluidic techniques, orthogonal CRISPR-Cas systems, lateral flow techniques, signal logic gates, and other technologies.
I.Combination of CRISPR-Cas System and Microfluidic Technology
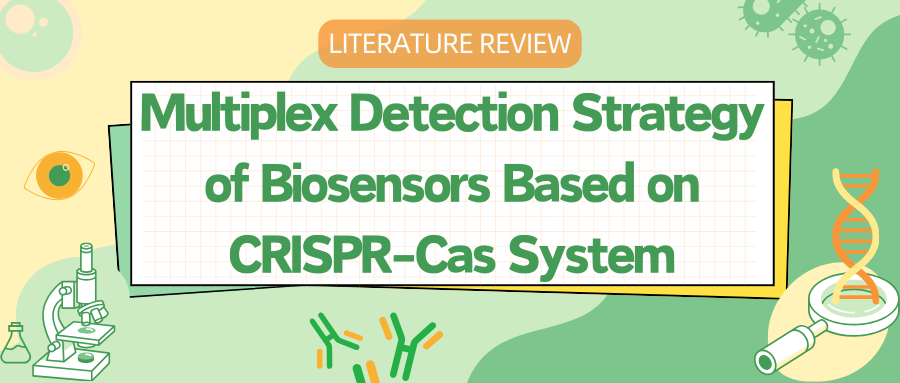
This strategy leverages microfluidic technology to precisely control and manipulate small fluid volumes in microarray wells, thereby resolving signal interference between targets and enabling the deployment of CRISPR/Cas systems for multiplex detection. Researchers have proposed the CARMEN assay, based on the CRISPR-Cas13 system, to achieve multiplex target detection. This method operates through the preparation of array chips with physically isolated micro-wells. Welch et al. developed mCARMEN based on the CARMEN assay, integrating commercial fluidic microfluidics and instrumentation to achieve multiplex detection with shorter detection times and simpler operating procedures. Despite the lightweight nature of the chip, its limitation lies in its single-use format, which increases relative detection costs, and it requires customized microscopes and chips, complicating clinical application procedures.
Similarly, utilizing microfluidic chip technology, a handheld microfluidic chip named CRISPR-RDB (CRISPR-Based Reverse Dot Blot) has been designed to detect four targets with a colorimetric output that is visible to the naked eye. This method not only realizes multiplex detection but also offers high sensitivity, high specificity, ease of operation, low cost, and obviates the need for large instruments, thereby providing opportunities for on-site detection. However, the membrane-labeled probes used in this technique cannot be stored for long periods and the number of detectable targets per test is limited. Xing et al. proposed a multiplex detection method integrating RAA technology with CRISPR-Cas12a and designed an easy-to-operate, long-term storage finger-actuated microfluidic biosensor (FA-MB). The RAA and CRISPR-Cas12a steps are mutually independent, and the required reagents can be lyophilized and embedded in the chip for long-term storage, achieving a breakthrough in portability.
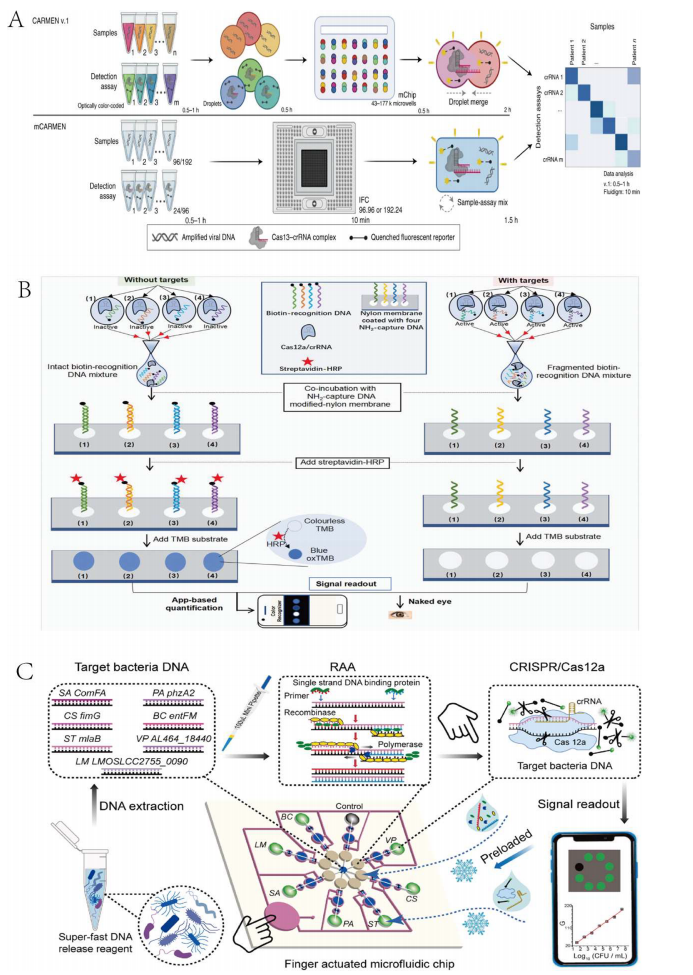
Figure 1 Multiplexed Target Detection Strategies Combining the CRISPR-Cas System with Microfluidic Technology
II. Orthogonal CRISPR-Cas System
Utilizing the substrate preferences of different Cas proteins and the orthogonality of different Cas enzymes is another feasible strategy for achieving multiplex detection based on the CRISPR-Cas system. By using various reporters or lateral flow strips, channel interference can be effectively eliminated.
Researchers have developed the CRISPR-TMSD platform, which utilizes the CRISPR-Cas12a system to detect multiple targets through toehold-mediated strand displacement (TMSD) reporters. Three TMSD reporters are cleaved by target-activated cis-cleavage, generating different fluorescence signals to achieve multiplex detection. However, this method suffers from limited sensitivity and instability. Jonathan S et al. developed the SHERLOCKv2 platform based on the specific cleavage preferences of different Cas enzymes for particular dinucleotide motifs, employing the orthogonality of LwaCas13a, PsmCas13b, CcaCas13b, and AsCas12a to detect four targets. Nevertheless, a drawback of this method is the need for separate RPA amplification of targets before detection. Using different Cas enzymes, Guk et al. proposed a mixed CRISPR-Cas system based on CRISPR orthogonality, mixing LbCas12a and LwCas13a and designing different crRNAs to separately detect DNA and RNA in a one-pot reaction.
While orthogonal CRISPR-Cas systems have achieved multiplex detection, several challenges remain, such as overlapping emission spectra of reporter genes and the variability in different Cas enzymes' preferences for reporter genes. These issues may limit the use of orthogonal enzymes and reporter genes. Signal interference might be resolved by physically separating the processes of signal generation and reception.
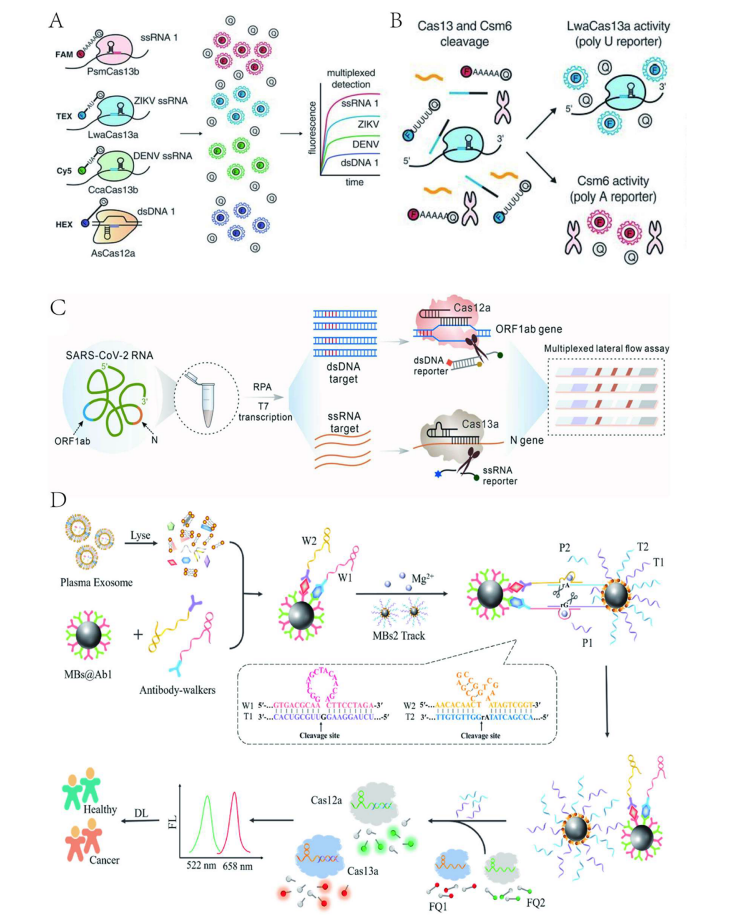
Figure 2 Multiplex Target Detection Strategy Based on Orthogonal CRISPR-Cas System
III. Lateral Flow Technology
Using lateral flow strips to address signal interference between targets is a common approach for achieving multiplex detection. For example, researchers have used CRISPR-Cas9 combined with biotin and digoxigenin to simultaneously detect two gene fragments of SARS-CoV-2 on lateral flow strips. This technique significantly improves detection accuracy and efficiency, making it suitable for POCT.
To apply multiplex detection platforms in practice, Yin et al. developed a self-contained paper-based lab platform for the multiplex genetic diagnosis of SARS-CoV-2. After target amplification in a 3D-printed RPA reactor, a sucrose valve opens, allowing targets to enter each detection chamber and activate CRISPR-Cas12a for multiplex detection. The platform has a detection time of about 1 hour and offers simple, low-cost, automated, multi-gene detection.
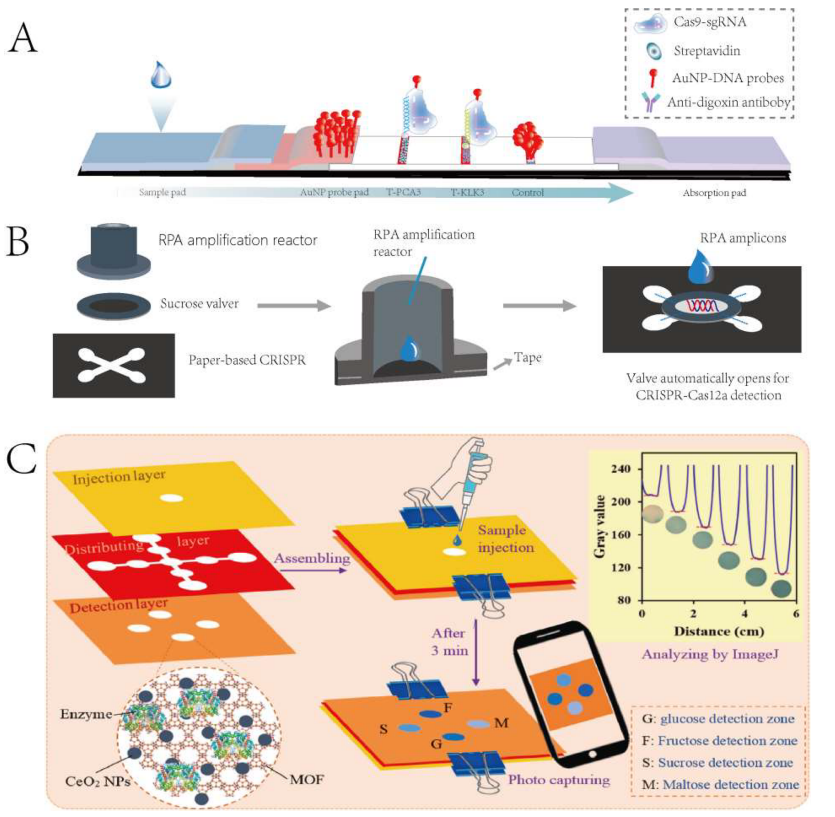
Figure 3 Multiplex Target Detection Strategy Based on Point-of-Care Testing (POCT) Platforms
IV. Signal Logic Gates
Logic gates perform fundamental logical functions and make decisions based on their inputs. Molecular logic gates constructed using CRISPR-Cas12a with different targets as inputs can achieve distinct signal responses, thereby avoiding signal interference and being applicable to established multiplex detection systems.
Gong et al. proposed a biosensing platform based on CRISPR-Cas12 integrated with a logic gate framework for the simultaneous detection of miR-944 and miR-205 expressed in lung cancer patients. When input miRNAs are present, the output signal of the AND logic gate is 1. This, in turn, triggers the release of DNA (target primers), resulting in a visible color change in the system. Conversely, in the absence of any target miRNA, the output of the logic gate is 0. This method cleverly enables the simultaneous detection of two targets, achieving a detection limit as low as 36.4 fM.
V. Combining CRISPR-Cas System with Other Technologies
Combining the CRISPR-Cas system with other technologies in the biosensor field can also form multiplex target detection strategies, including PhC barcodes, quantum dots, and DNA tetrahedrons.
Researchers proposed a CRISPR-Cas9 multiplex detection platform based on bio-inspired photonic crystal (PhC) barcodes. Different probes combined with structurally colored barcodes of unique wavelengths allow different PhC barcodes to capture and cleave target sequences recognized by the CRISPR-Cas9 system. Although this method has high sensitivity, it is time-consuming and complex to operate.
Using high-luminescence quantum dots, Green et al. designed a nucleic acid hairpin (QDHP) molecular beacon for the quantitative detection of nucleic acids using CRISPR-Cas. The combination of QDs with CRISPR-based detection broadens its application scope.
Zhan et al. proposed a CRISPR biosensor based on DNA tetrahedrons (DNT) for the simultaneous detection of multiple HR-HPV-DNA, further improving sensitivity and reducing detection time.
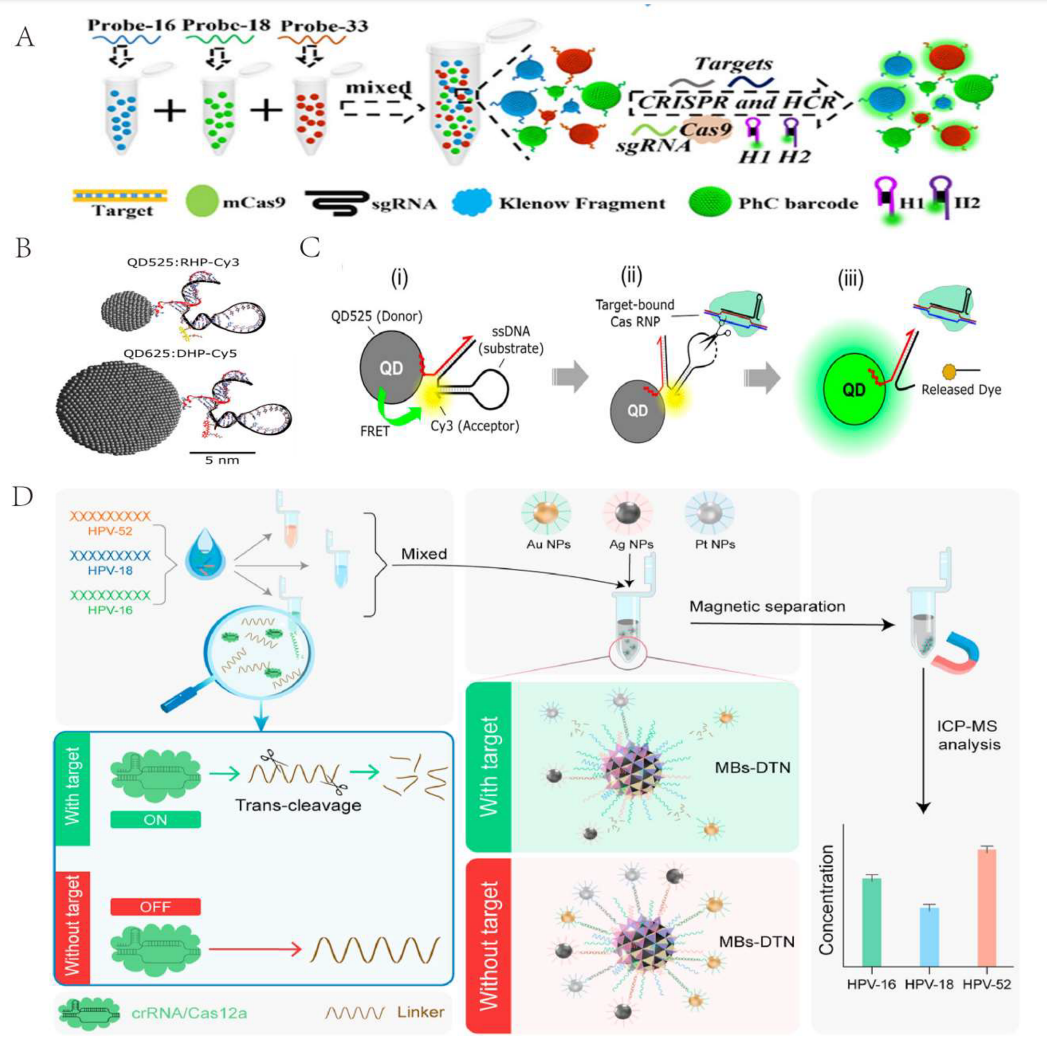
Figure 4 Multiplex Target Detection Strategies Combining CRISPR-Cas System with Other Technologies
VI. Detection of other multiplexes
This strategy encompasses multiplex detection based on single-pot CRISPR technology and biosensor-based multiplex detection. Single-pot CRISPR multiplexing poses challenges due to non-specific amplification caused by multiple primers and the indistinguishable trans-cleavage activity of Cas proteins, often necessitating the integration of additional technologies to achieve multiplex detection. In response, Shang et al. designed a multiplex detection platform utilizing CRISPR-Cas13a and microfluidic droplets for the detection of multiple bacterial species, ultimately obtaining results through digital droplet readings. A limitation of this platform is the requirement for droplet chips and specialized operation, making it difficult to apply in point-of-care testing.
In biosensor-based multiplex detection, distinguishing the fluorescent signals produced by multiple targets remains a challenge. Johnston et al. proposed a multi-channel biosensor, BiosensorX, that integrates electrochemical technology to sequentially arrange different detection methods on a single-channel microfluidic chip, allowing for the simultaneous detection of SARS-CoV-2 variants, suitable for point-of-care testing.
In summary, implementing CRISPR-Cas system-based multiplex detection strategies within a single sample can enhance detection efficiency and facilitate disease diagnosis. The first important strategy for achieving multiplex detection is the integration with microfluidic technology, which can generate separate spatial detection for different targets with the same signal. The second strategy involves the orthogonal reactions of different Cas enzymes, enabling various Cas proteins to recognize distinct target substances. Furthermore, new materials and innovative strategies can be developed to facilitate the simultaneous detection of multiple targets with the CRISPR-Cas system. The integration of CRISPR-based multiplex detection with other technologies or platforms holds significant potential for point-of-care testing (POCT).
Additionally, CRISPR-Cas system-mediated biosensor multiplex detection still presents several avenues for development. For example, researchers could combine novel materials such as quantum dots to create new biosensing platforms for immediate care testing or develop special strategies for multiplex detection of the CRISPR-Cas system through designs like DNA dumbbell probes.
EDITGENE boasts its independently developed protein purification platform. Our Cas enzymes have shown significantly superior activity compared to NEB and other market enzymes through active testing, achieving the highest standards in purity, activity, and sensitivity.
Be on the look out for our isothermal amplification kit and CRISPR detection kits, featuring low equipment requirements and short reaction times—Coming soon!
Recent Blogs:
1.[Literature Review] Key Molecular Mechanisms of Prime Editing Revealed by Cryo-Electron Microscopy
2.[Star of the Month] Human Genome KO Library,Mouse Kinome CRISPR KO Library,Yeo Lab RNA-binding Protein CRISPR KO Library
3.[Literature Review] The CRISPRi Screen Helps Find Key Genes that Regulate Glioblastoma
Follow us on social media
联系我们 / Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






