[Literature Review] The CRISPRi Screen Helps Find Key Genes that Regulate Glioblastoma

In a recent publication in PLOS GENETICS, researchers at the Keck School of Medicine of the University of Southern California (USC) presented the results of a CRISPRi screen of long non-coding RNAs regulating glioblastoma. The study, entitled "CRISPRi screen of long non-coding RNAs identifies LINC03045 regulating glioblastoma invasion," highlights the potential of long non-coding RNAs as novel targets for invasive studies in glioblastoma (GBM). Nevertheless, long non-coding RNAs (lncRNAs) have recently emerged as promising new targets for gene therapy due to their high degree of cell type-specific expression and function. In this study, researchers employed CRISPRi (CRISPR interference) technology to conduct an lncRNA loss-of-function screen to assess the invasive function of GBM-associated lncRNAs. This approach, coupled with mechanistic characterization and candidate gene analysis, ultimately led to the identification of LINC03045 as a pivotal non-coding gene in glioblastoma invasion. This novel lncRNA may regulate invasion through WASF3, and further characterization of this gene is anticipated to establish it as a potential therapeutic target for future glioblastoma treatment.
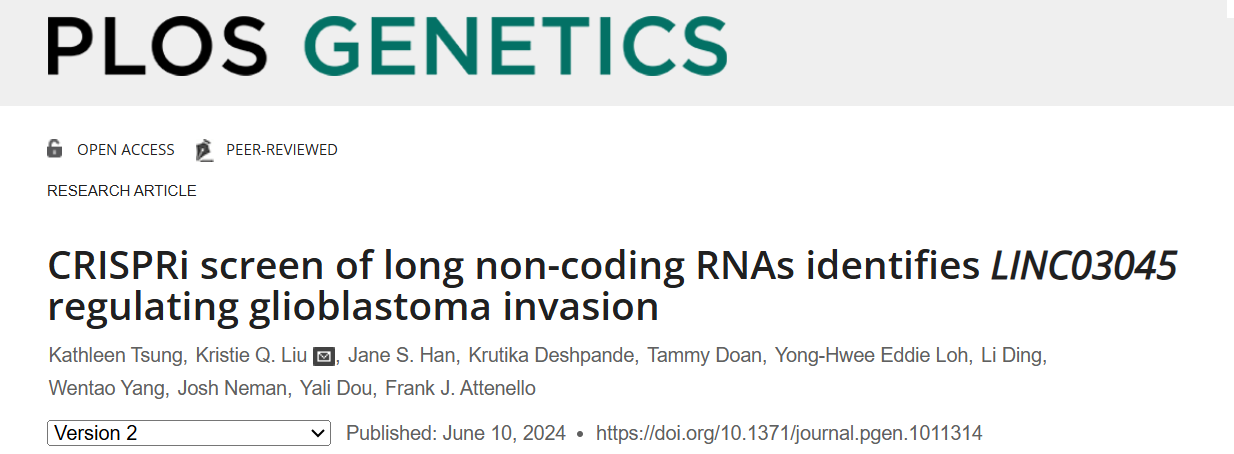
Glioblastoma (GBM) is the most prevalent and aggressive primary malignant brain tumor, with a high annual incidence. The established targeted therapies face significant challenges, including tumor recurrence and increased resistance to chemotherapy and radiation. To address these shortcomings, novel therapeutic strategies and molecular targets have emerged as a crucial area of research. To date, few studies have specifically identified novel candidate long non-coding RNAs (lncRNAs) against gliomas. In response, the clustered regularly interspaced short palindromic repeats (CRISPR) interference (i) technology enables unbiased invasive screening. By employing CRISPRi and oligonucleotide (ASO) knockouts, researchers were able to validate the top-screened candidate genes in three tumor lines. The clinical relevance of the candidate genes was evaluated through analyses conducted by The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) on survival outcomes. The mediators of lncRNA effects were identified through differential lncRNA expression analyses of the knockouts. Finally, tumor invasiveness was assessed through knockout and rescue experiments.
I.CRISPRi Screening Can Identify IncRNAs Involved in Invasion
To identify and characterize long non-coding RNAs (lncRNAs) that regulate glioma cell invasion in vitro, the researchers utilized CRISPR interference (CRISPRi) for gene knockout. They conducted a screening process involving a U87-specific CRISPRi library packaged into a lentivirus, which was then transduced into U87 cells stably expressing dCas9-KRAB. Following this, the infected cells were subjected to puromycin screening and subsequently inoculated into a Matrigel Boyden chamber. Subsequently, DNA was extracted from the screened cells and subjected to sequencing analysis.
Researchers repeated the CRISPRi screening process, averaged the screening replicates, and used the first 3 sgRNAs of each lncRNA (Mann-Whitney P-value) to determine the screen hits compared to the non-targeted controls. This screening score is plotted via a volcano plot, where the dashed line indicates the threshold for determining screen hits. In non-invasive cells, 45 gene targets were significantly enriched in the library, representing genes essential for glioma cell invasion. Ultimately the researchers identified positive hits through this process and the most suitable candidate was determined to be LINC03045 (annotation LH02236) on chromosome 10.
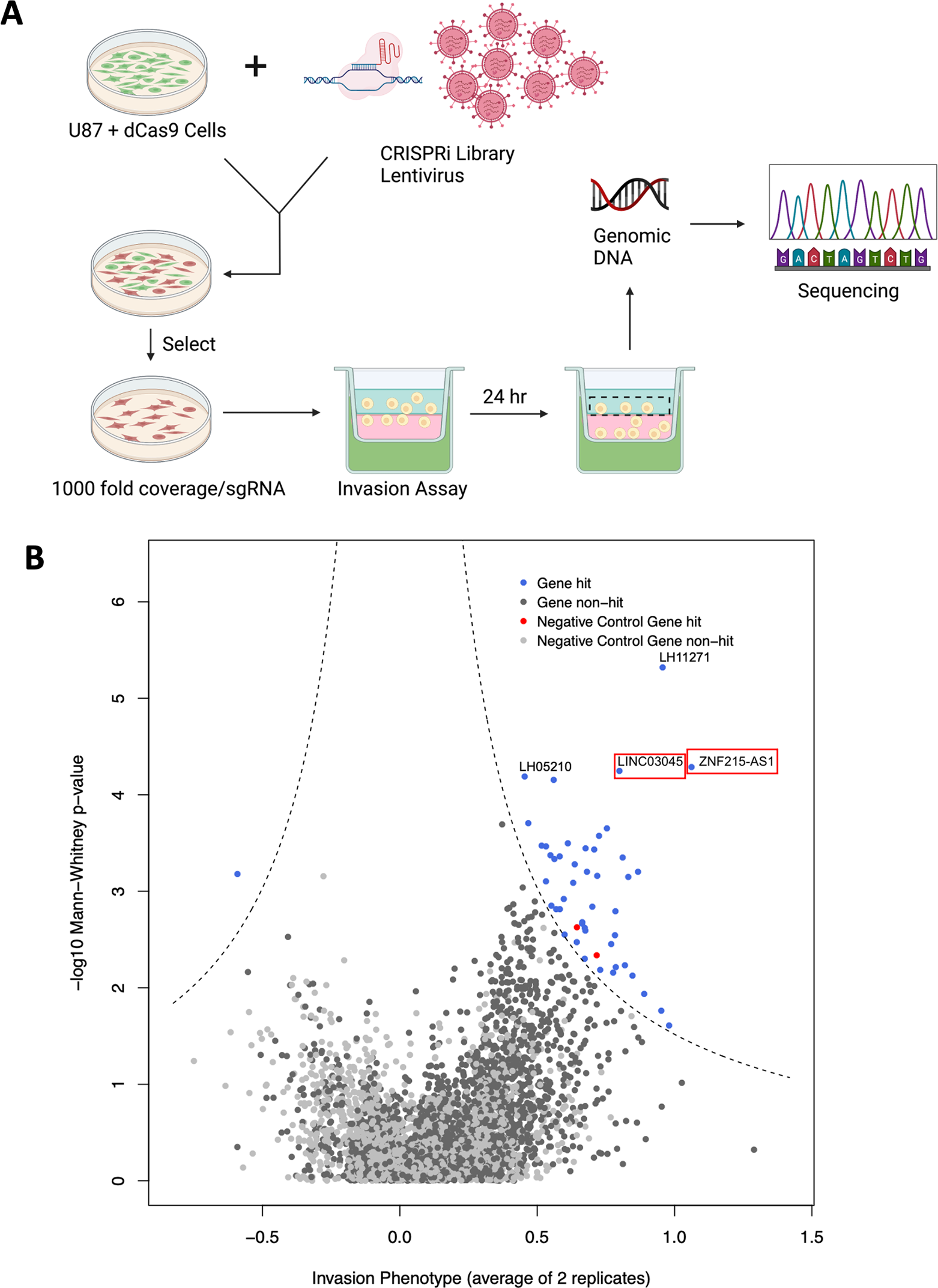
Figure 1: Results of the CRISPRi Screening Programme
To better interpret LINC03045, the researchers measured in counts per million (CPM) and used positional coordinates to assess LINC03045 expression levels. Both glioblastoma GBM (3.6 ± 0.27 CPM) and low-grade glioma LGG (2.06 ± 0.30 CPM) had higher mean LINC03045 gene expression than normal cerebral cortex (0.67 ± 0.06 CPM) (mean ± SEM). When comparing tumors and controls, LINC03045 showed significant expression changes.
Researchers performed a Kaplan-Meier (KM) survival analysis of all patients with glioma (stage 2-4) using LINC03045 counts. Cohorts divided into quartiles showed a significant decrease in survival with increasing expression levels (p<0.0001). In addition, Cox regression analysis of the cohort showed that 2.5 LINC03045 CPM could be divided into high-risk and low-risk groups. Survival was significantly lower (p<0001) in the high risk group (>2.5 CPM). The hazard ratio (HR) from 0 to 2.5 CPM was 1.99 for each additional CPM.
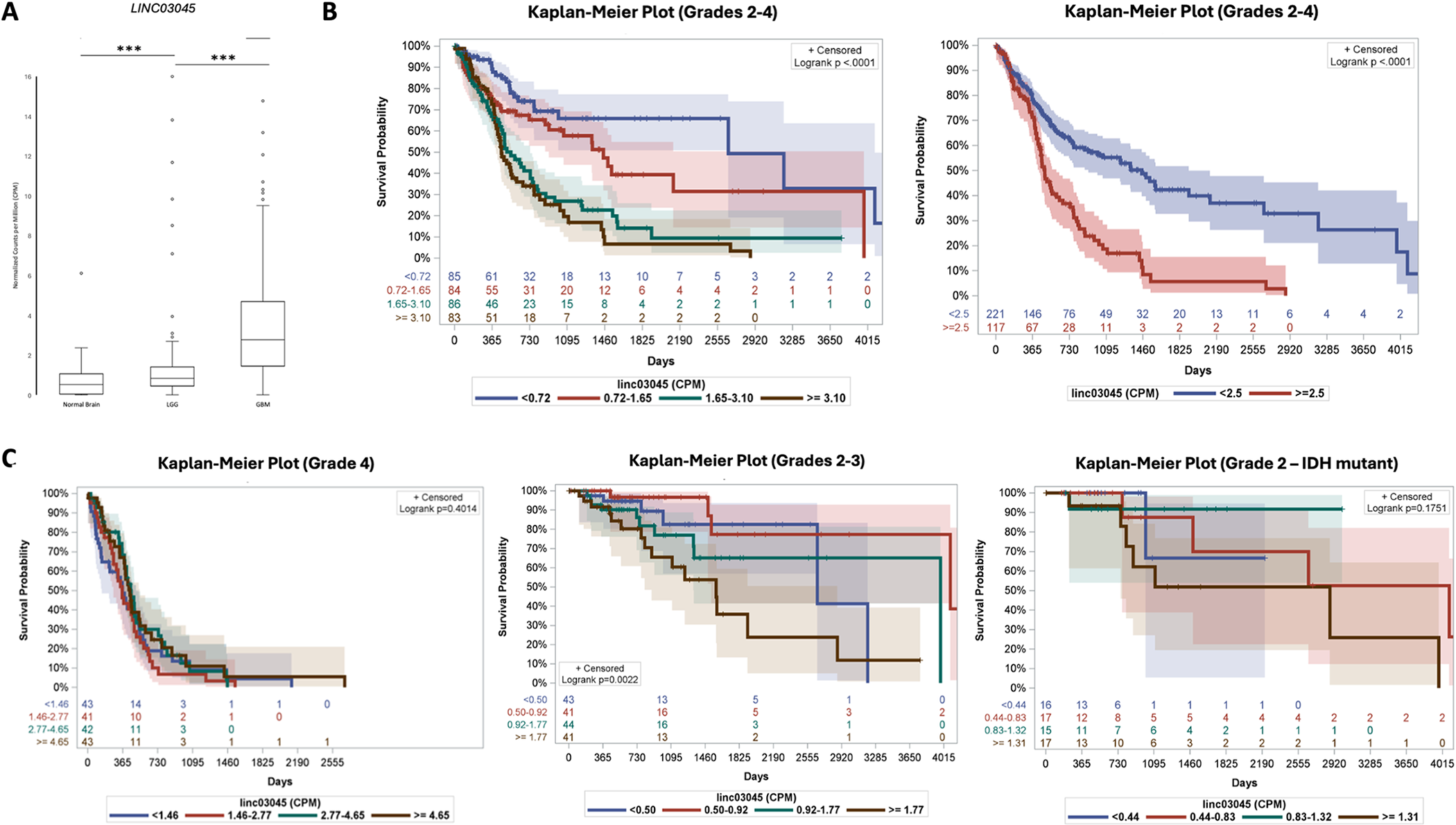
Figure 2: Expression of LINC03045 in Patient Glioma Samples from The Cancer Genome Atlas (TCGA) and Genotypic Tissue Expression (GTEx) Projects
III.Single sgRNA Knockout Enables Repeated Screening of Identified lncRNA Phenotypes
To confirm whether the screened candidate genes exhibited consistent phenotypes following single lncRNA inhibition, the researchers conducted CRISPRi knockout experiments using the first two sgRNAs of each candidate gene. The results demonstrated that LINC03045 exhibited the most pronounced effect size and reproducibility in both post-knockout transcriptional expression and invasion assays.
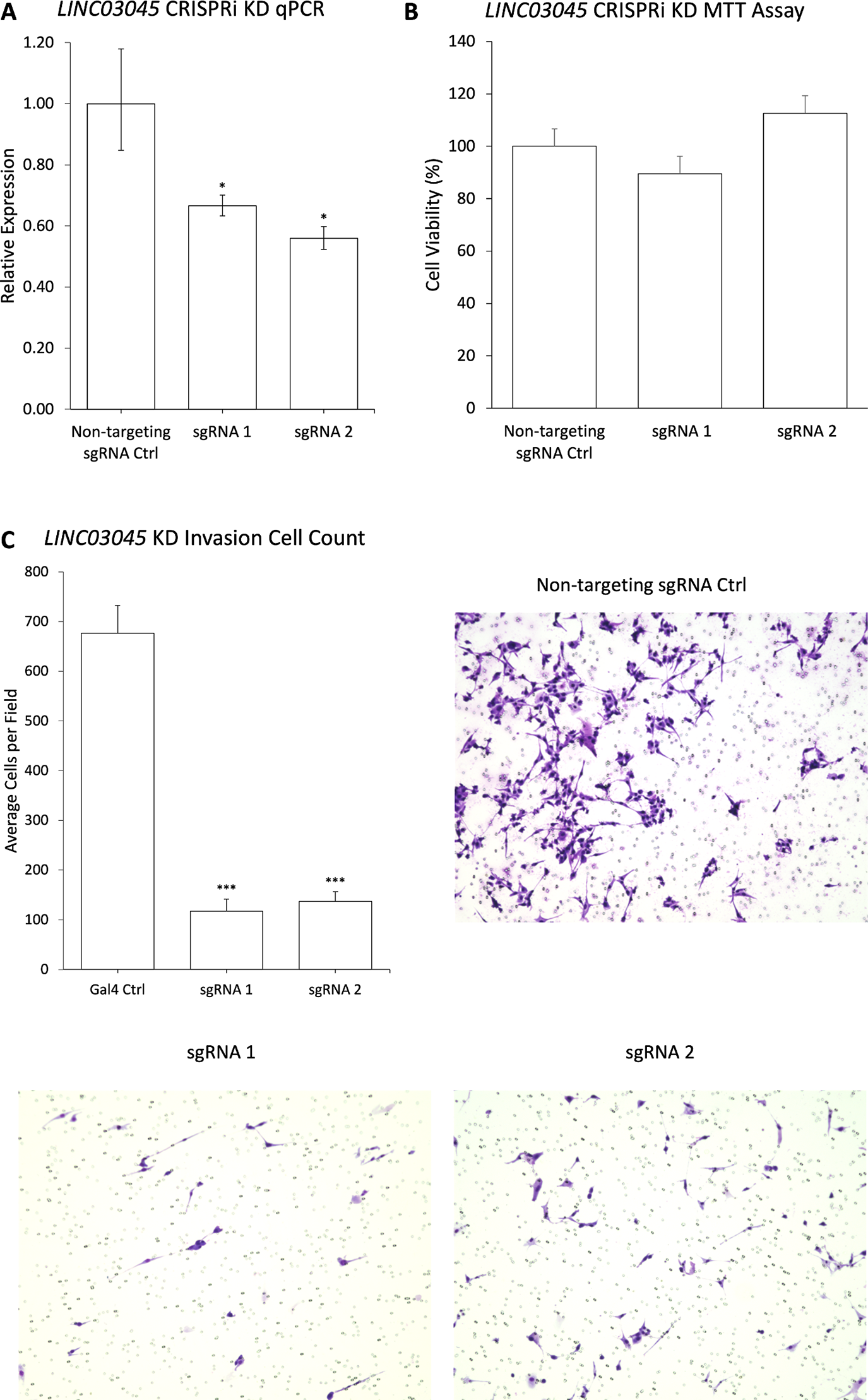
Figure 3: Validation of CRISPRi Screening Results
IV.Antisense Oligonucleotide-mediated KD of LINC03045 is Consistent with CRISPRi Results in Multiple Cell Lines
To validate the functional screening accuracy of the CRISPRi knockout technology in this study and to assess the potential of this LINC03045 knockout technology to reduce malignant glioma cell invasion and transformation using pharmacological approaches, the investigators employed antisense oligonucleotide-mediated knockout as a second KD approach to confirm its application in three glioma cell lines: the patient tumor line USC02, the commercial tumor lines U87 and U251. The U251 glioma cell line was also utilized. As a consequence, qPCR transcriptional expression following ASO KD demonstrated a reduction in transcriptional expression. The MTT assay indicated that there was no change in proliferation following ASO KD of LINC03045 in USC02, U87, and U251 cells. Conversely, in vitro invasion, as evaluated by Matrigel assay following ASO-mediated KD of LINC03045, exhibited a decline in invasion in USC02, U87, and U251, accompanied by a reduction in invasion. The collective findings suggest that antisense oligonucleotide-mediated knockout of LINC03045 yields comparable outcomes to CRISPRi in a range of cellular models.
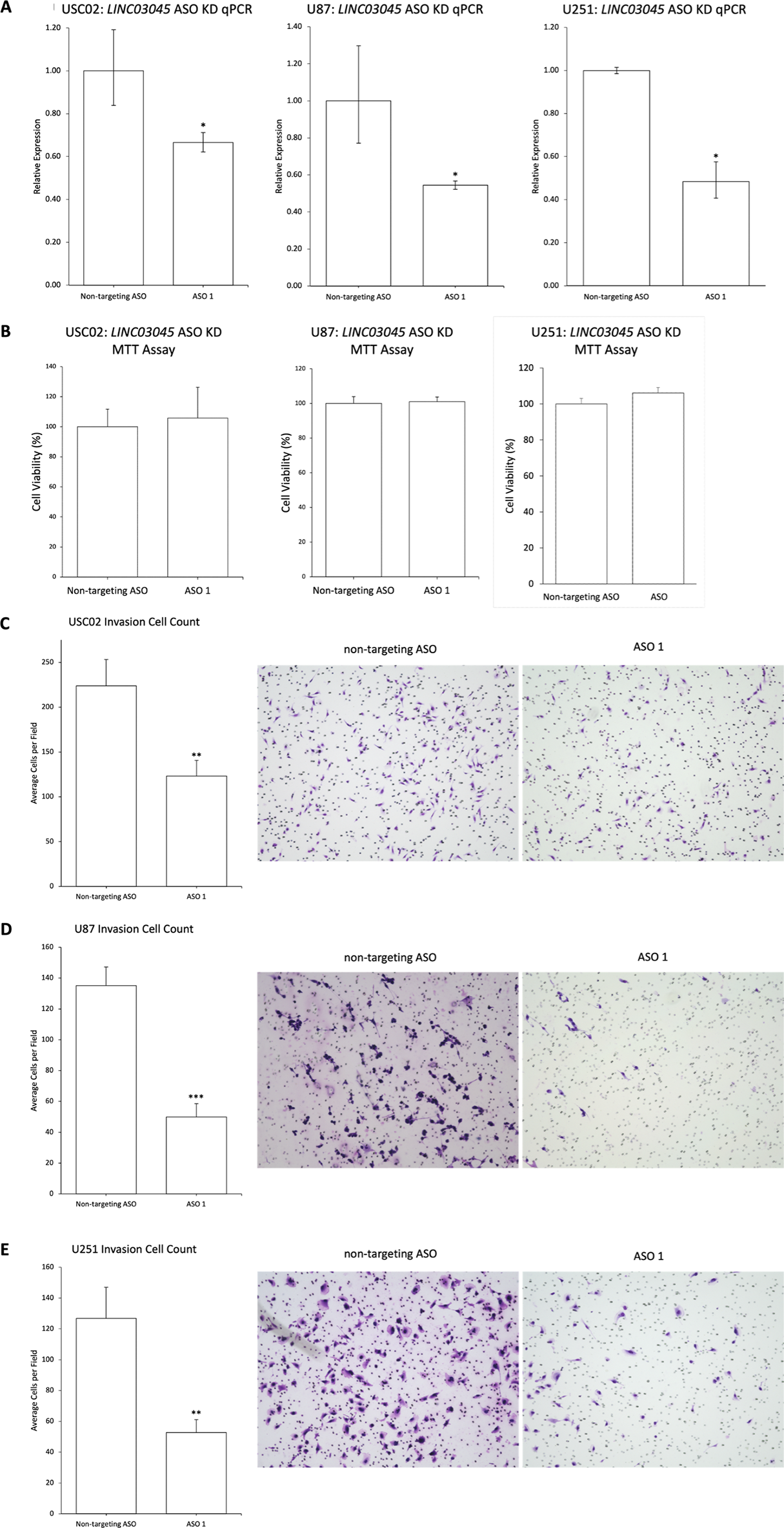
Figure 4: Antisense Oligonucleotide-mediated KD Replication CRISPRi Results for LINC03045 in Multiple Cell Lines
V.Genome-wide Differential Gene Expression by TCGA and GTEx Analysis
To understand the association of LINC03045 with genome-wide gene expression changes, the researchers further explored the TCGA and GTEx patient data. First, they used LINC03045 expression distribution analysis to clarify that there was a clear difference between the top 25% of patients and the bottom 25% of patients. Subsequently, they divided TCGA-GBM patients into two groups based on LINC03045 expression levels. Evaluation of the cohort of patients with low LINC03045 expression revealed significant overall alterations in gene expression, as demonstrated in the top 100 and 500 genes with the most alterations associated with low lncRNA expression. KEGG pathway analysis revealed that LINC03045 expression was significantly correlated with oxidative phosphorylation as well as ribosome and spliceosome activity. Gene ontology analysis revealed a broad association between RNA transcription, RNA polymerase II activity, RNA splicing and chromatin organization in the increased expression of LINC03045. Among the gene ontology pathways involved, WASF3 was significantly associated with the enrichment pathway involved in cytoskeletal regulation (GO:0007010).
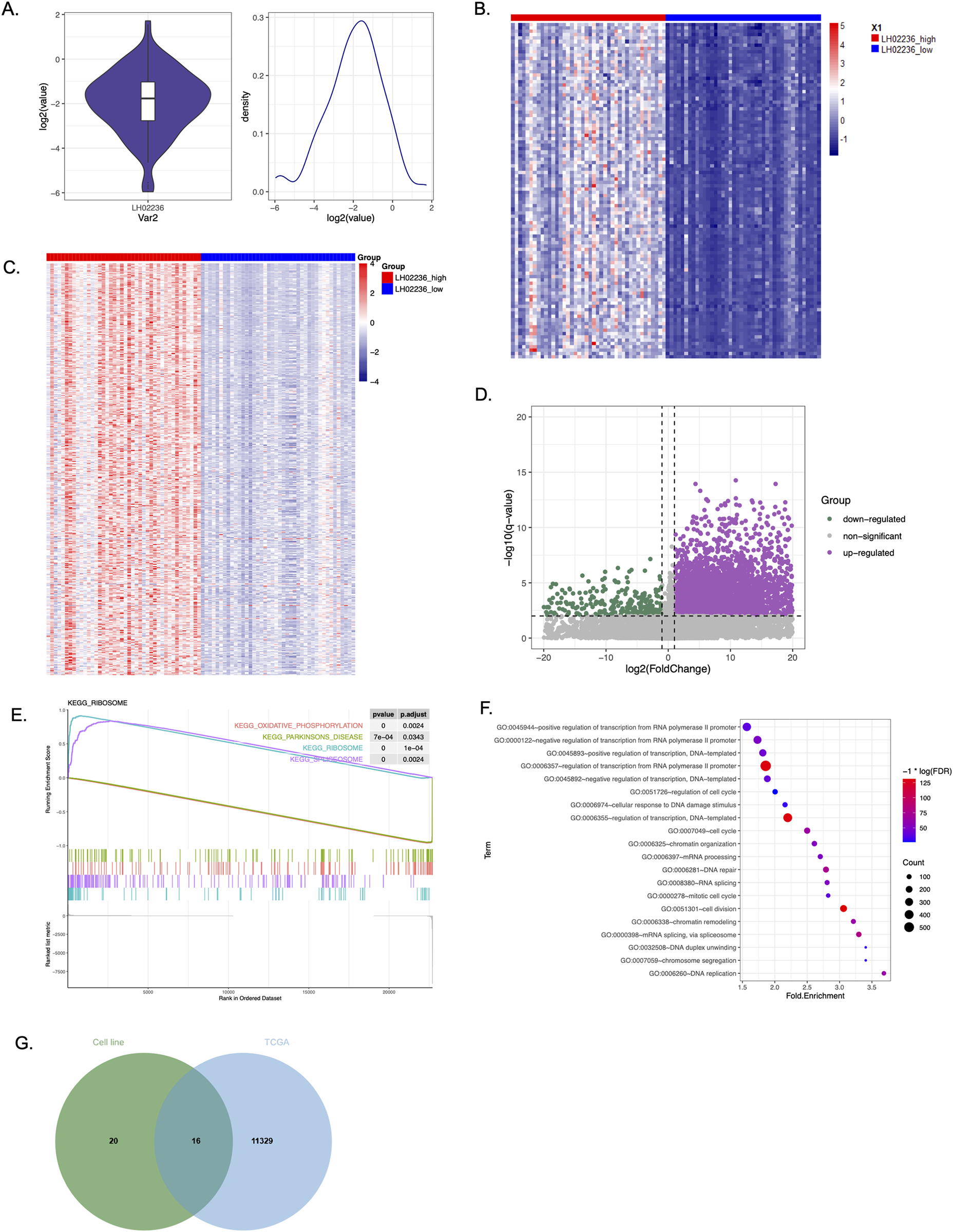
Figure 5: Genome-wide Differential Gene Expression from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEx) projects
VI.WASF3 is the Downstream Medium for LINC03045.
To investigate the downstream effects of LINC03045 and to clarify the potential mechanisms mediating LINC03045 activity, the researchers performed ASO-mediated KD of LINC03045 using U87 cells to assess gene expression by RNAseq.The results of the RNAseq analysis indicated that a number of genes exhibited a notable increase or decrease in expression following the knockout of LINC03045. A comparison of the candidate genes to gene data related to LINC03045 expression in TCGA patients revealed a significant positive correlation between the expression of the WASF3 gene and that of LINC03045. Furthermore, WASF3 was identified as the only candidate gene significantly downregulated in cohorts of patients with low LINC03045 expression. To ascertain the impact of WASF3 knockout on cellular invasion, the researchers conducted a matrix analysis and a three-dimensional invasion assay. The results demonstrated that siRNA-mediated invasion of the surrounding matrix was diminished in WASF3 globules relative to untransfected U87 globules. Furthermore, a statistically significant positive correlation was observed between WASF3 and LINC03045 (p=0.016). Furthermore, the relationship between LINC03045 and WASF3 was confirmed by rescuing the invasive ability of the patient-derived cell line USC02 after LINC03045 KD through the overexpression of ASF3 in an invasiveness assay. The WASF3 rescue assay provided confirmation that WASF3 overexpression restored the invasive loss caused by lncRNA KD. The comprehensive analysis demonstrated that WASF3 is a mediator of the tumor effects of LINC03045.
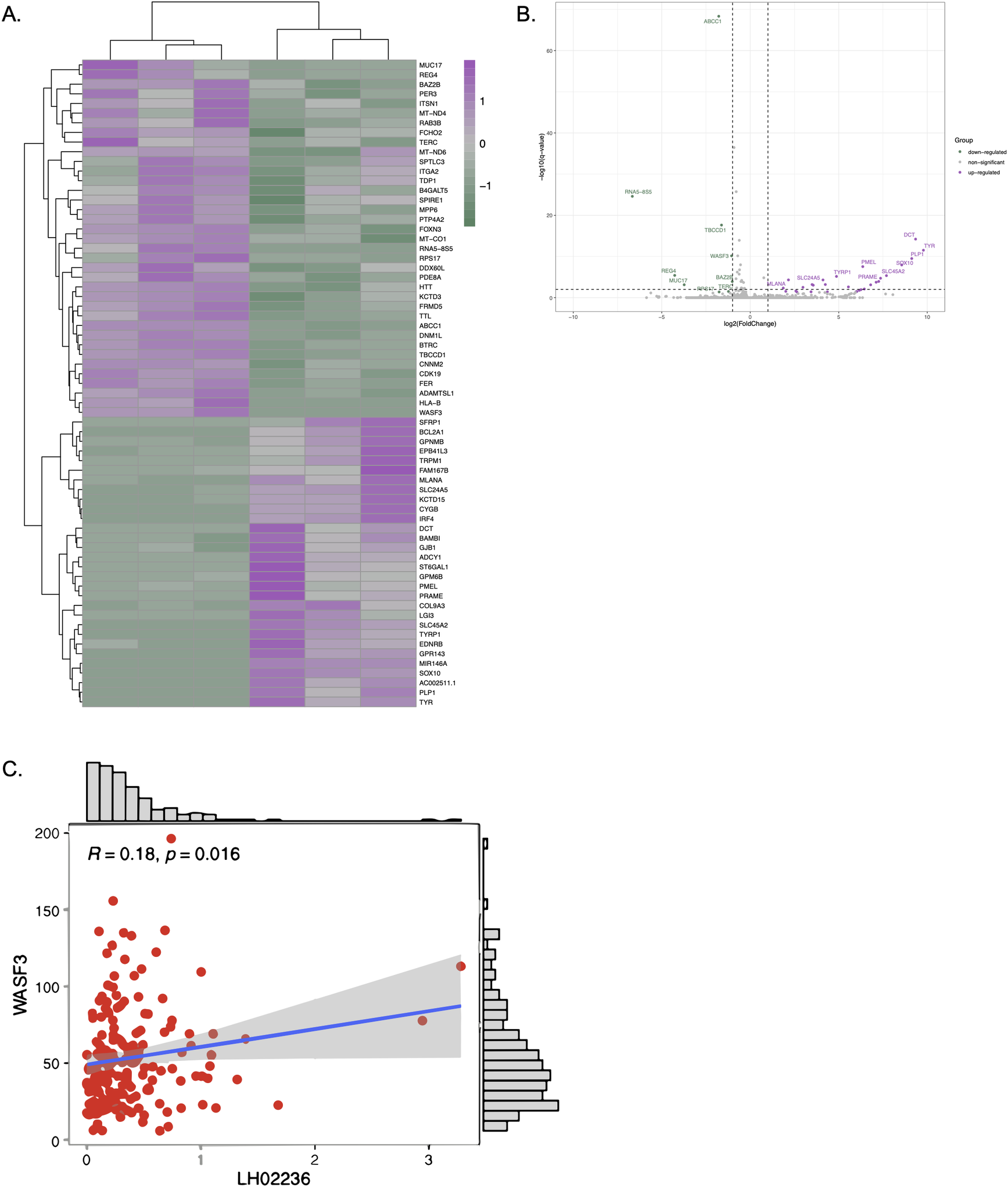
Figure 6: RNAseq Analysis of LINC03045 KD Cells
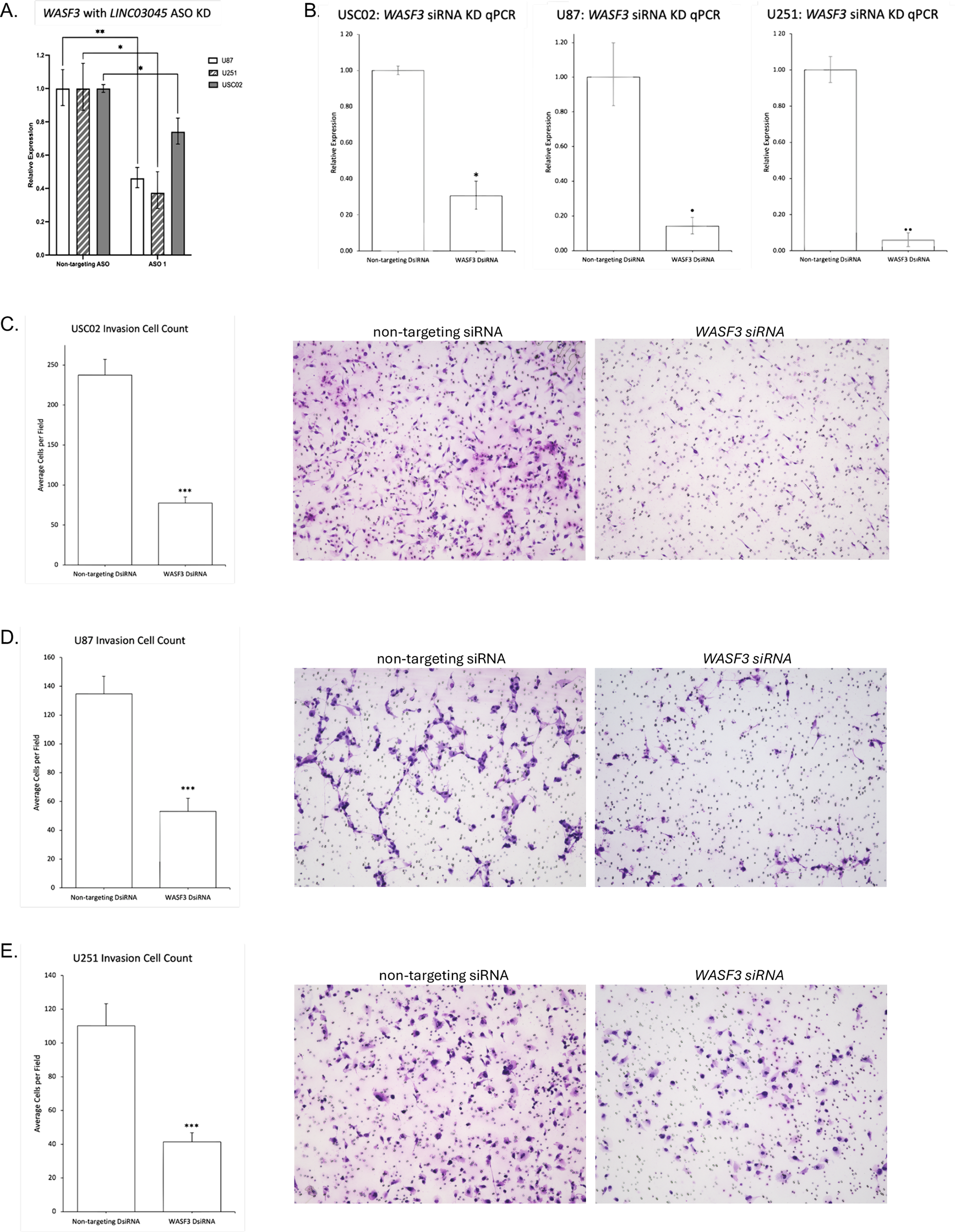
Figure 7 WASF3 Impact Encroachment
In conclusion, the researcher conducted a functional screen utilizing large-scale CRISPRi and identified LINC03045 as a pivotal non-coding gene in glioblastoma invasion. Concurrently, RNA-seq and mechanistic studies indicated that this novel lncRNA may regulate invasion through WASF3, which could offer new therapeutic targets for future glioblastoma treatment.
EDITGENE offers a complete solution for CRISPR library screening,CRISPR KO/CRISPRa/CRISPRi, and the most extensive collection of library plasmids/viruses, which is available within weeks.
Recent Blogs:
2.【Cutting-Edge News】New Trends in Gene Editing - FGFR1, BRD4, and BACE1 Knockout Cells
3.[Cutting-Edge News] Advances in Prime Editing: Three Strategies to Enhance Editing Efficiency
Follow us on social media






