【Cutting-Edge News】Developments in Gene Editing - fut8, atm, and brca1 Offers New Perspectives for Cancer Treatment

Gene knockout cells refer to cells that have undergone gene editing using CRISPR technology. This involves designing specific sgRNAs to target the cleavage site of the gene, allowing the Cas9 protein to bind to the sgRNA and “cut” the target gene out of the cell, thus achieving gene knockout. These cells hold vast potential in various fields such as life sciences, medicine, and drug development. Applications include creating disease models, conducting drug screening and target validation, and achieving gene therapy by knocking out disease-causing genes. In this article, we select three studies on gene knockout cells to interpret their findings and bring you up to date on the latest advancements in this cutting-edge technology.
I. FUT8-KO Cells Reveal New Therapeutic Targets for Pancreatic Cancer Treatment
Original Link: <https://doi.org/10.1016/j.bbagen.2021.129870>
Pancreatic ductal adenocarcinoma (PDAC), accounting for 90% of all pancreatic cancers, is an extremely malignant tumor. Aberrant glycosylation changes on cancer cell surfaces have been shown to correlate positively with tumor progression and metastasis. α1,6-Fucosyltransferase 8 (FUT8), the key enzyme responsible for catalyzing core fucosylation, is abnormally expressed and activated in various malignancies, implicating its involvement in multiple physiological and pathological processes. Although FUT8's role has been observed in other cancer types, its specific molecular mechanisms in PDAC's malignant transformation and potential as a therapeutic target remain unclear.
Researchers employed the CRISPR/Cas9 system to knock out the FUT8 gene in MIA PaCa-2 and PANC-1 cell lines. They assessed the migratory ability of FUT8-KO cells using Transwell migration and wound healing assays, and their proliferative capacity using MTT assays and colony formation experiments. Additionally, they examined the expression levels of cancer stem cell markers in FUT8-KO cells. The results demonstrated that FUT8-KO cells exhibited significantly reduced migration, proliferation, and colony formation abilities compared to wild-type cells, along with decreased expression of cancer stem cell markers. The study using FUT8-knockout cells highlights the crucial role of FUT8 in pancreatic cancer and suggests that FUT8 could be a potential therapeutic target for PDAC, offering a novel perspective for future treatment strategies.
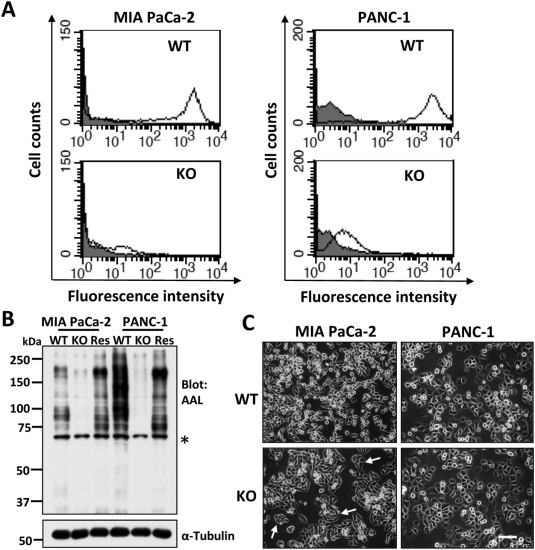
Figure 1: Generation of FUT8-KO Cells
II. ATM Gene Knockout Leads to Proteasomal Degradation of FANCD2 Protein
Original Link: <https://doi.org/10.1186/s12885-023-10772-y>
Neuroblastoma (NB) is a common pediatric solid tumor with high heterogeneity in clinical presentation and prognosis. Despite various treatment strategies, high-risk NB patients often exhibit resistance to standard therapies and may develop metastasis. The ATM gene is associated with DNA damage response, and its heterozygous loss on chromosome 11q and hemizygous mutations of the ATM gene are mutually exclusive in NB tumors. ATM knockdown has been shown to promote tumor formation in vitro and in vivo in NB cell lines, but the relationship between ATM and tumorigenesis or cancer invasiveness is not fully understood.
Researchers employed CRISPR/Cas9 technology to knock out the ATM gene in NGP and CHP-134 NB cell lines. They analyzed the proliferation and colony formation abilities of ATM-knockout cells, assessed the expression of various proteins involved in DNA repair pathways by Western blot, and stably transfected FANCD2 expression plasmids into ATM-knockout cells to overexpress FANCD2, followed by immunofluorescence microscopy to determine protein expression. The results showed that ATM deficiency led to reduced FANCD2 protein levels, and ATM-knockout cells exhibited increased sensitivity to the PARP inhibitor olaparib. Reintroduction of FANCD2 expression in ATM-knockout cells restored their proliferative capacity. ATM-knockout cells elucidate the mechanisms of ATM heterozygosity in neuroblastoma and demonstrate how ATM inactivation enhances NB cell sensitivity to olaparib treatment, which holds significant implications for high-risk NB patients with ATM dose-related and invasive cancer progression.
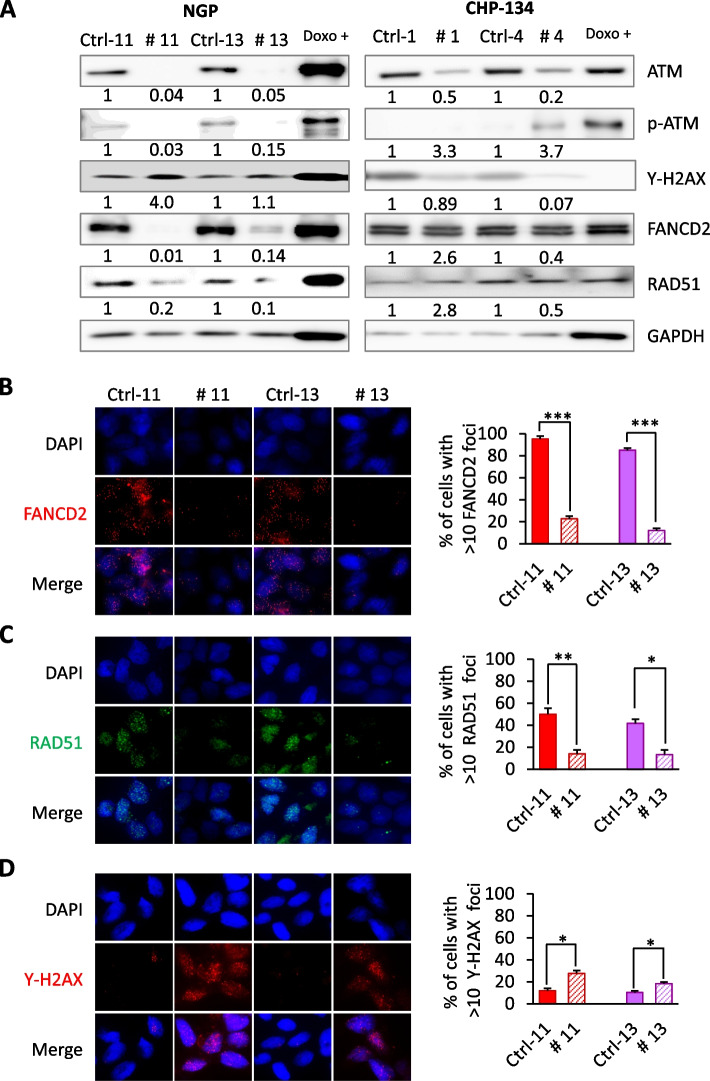
Figure 2: ATM Gene Deletion Impairs FANCD2 Expression
III. BRCA1 Gene Regulates HMGA2 Levels in Swan71 Trophoblast Cells
Original Link: <https://doi.org/10.1002/mrd.23255>
During early placental development, tumor suppressor genes and oncogenes work together to regulate cell proliferation and differentiation in a controlled manner. BRCA1 is a known tumor suppressor gene that forms a complex with ZNF350 and CtIP, binding to the promoter region of the HMGA2 gene and preventing its transcription. This regulation has been studied in cancer cells but less so in placental cells.
Researchers employed the CRISPR-Cas9 system to knock out the BRCA1 gene in the Swan71 cell line, generating BRCA1 knockout (BRCA1 KO) cells. They also overexpressed miR-182 in these cells by infecting them with lentiviral particles containing a miR-182 overexpression construct. The results showed that BRCA1 KO cells exhibited significantly increased levels of HMGA2 mRNA and protein compared to wild-type cells. Additionally, the overexpression of miR-182 led to a decrease in BRCA1 protein levels and an increase in HMGA2 protein levels. The study using BRCA1 knockout cells indicates that BRCA1 plays a crucial role in regulating HMGA2 levels in trophoblast cells and may be involved in placental development and function by affecting cell apoptosis. This provides a new perspective for understanding the role of BRCA1 in placental development.
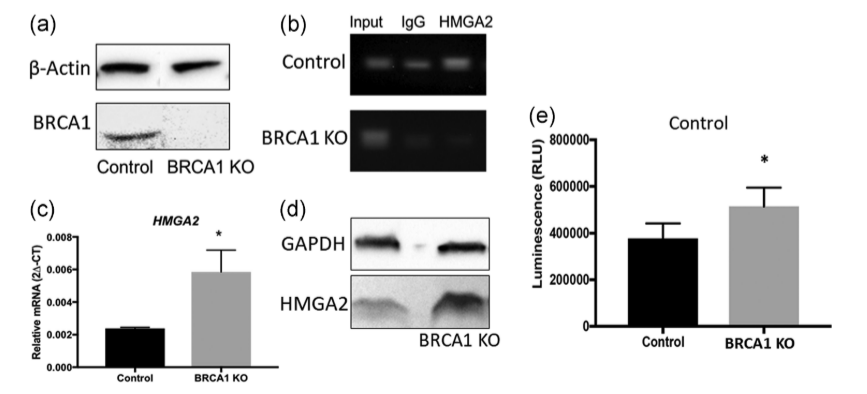
Figure 3: Expression of HMGA2 in Swan71 BRCA1 KO Cells
EDITGENE offers over 3800+ ready-to-use gene knockout cell lines, covering popular targets such as fut8, atm, and brca1 mentioned in the articles above. We provide one-stop scientific research support. Order now and receive your cells within weeks!
Recent Blogs
1.[Cutting-Edge News] Advances in Prime Editing: Three Strategies to Enhance Editing Efficiency
2.[Literature Review] Exploring the Functional Conservation of Cross-Species lncRNAs Using Cas12a Knockout Technology
3.[Frontier Information] CRISPRi Screening Boosts the Discovery of Key Targets in Tumor
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






