[Cutting-Edge News] Advances in Prime Editing: Three Strategies to Enhance Editing Efficiency

Prime editing, an advanced genome editing technology derived from the CRISPR/Cas system, facilitates precise insertion, deletion, and arbitrary base substitution at targeted genomic sites, offering higher accuracy and lower off-target rates. Since its introduction, it has attracted significant interest from the scientific community and is widely employed in researching genetic disorders, mutation-driven diseases, and cancer treatments. For instance, it can be used to correct disease-causing mutations in patient cells for treating genetic diseases or to target oncogenes in cancer cells to inhibit their growth and spread.
Original Article Link: <https://doi.org/10.1016/j.ymthe.2022.07.001>
Researchers screened 11 distinct RT variants, optimized their human codon usage with GenScript algorithms, resulting in a 1.4-fold increase in PE protein expression. By removing the RNase H domain and further shortening the RT sequence, truncated PECO variants were generated, reducing the Prime editor's length by 621bp without compromising editing efficiency. To facilitate dual AAV delivery of PE, they constructed split PE systems based on various Cas9 cleavage sites and inteins, identifying the Rma 573-574 and 674-675 cleavage sites, which, in conjunction with the Rma intein, significantly improved AAV titers and Prime editor efficiency. Through engineering modifications and optimization, the researchers successfully enhanced Prime editing efficiency and addressed the limitations imposed by AAV vector size, providing a more efficient and practical tool for future gene therapy and biomedical studies.
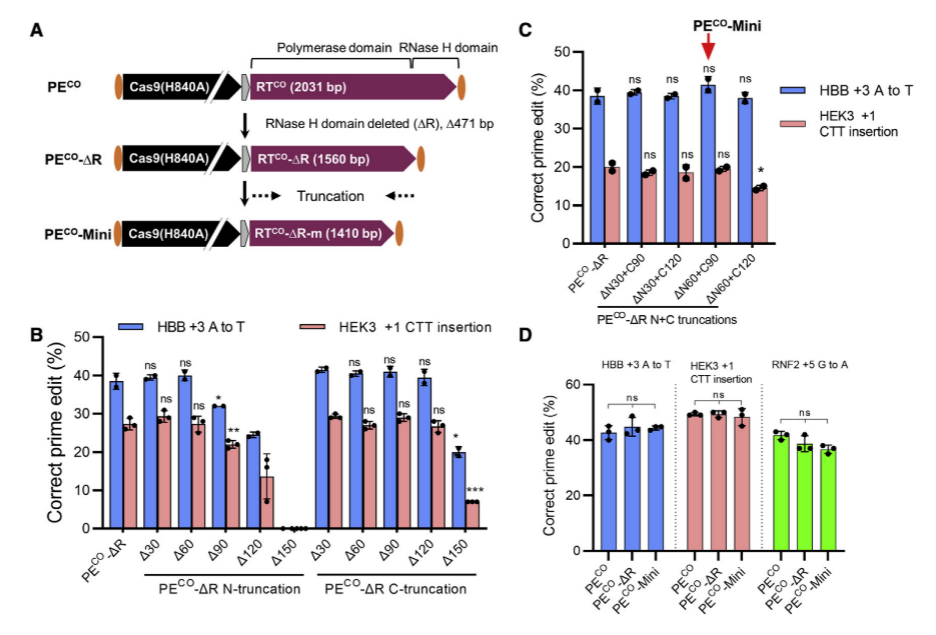
Figure 1: Optimizing Prime Editor Length while Maintaining Editing Efficiency
Original Article Link: <https://doi.org/10.1038/s41586-024-07497-8>
The molecular mechanism by which the Prime editor recognizes pegRNA and interacts with target DNA remains unclear, limiting the understanding and further optimization of the Prime editing process. To address this issue, researchers determined the cryo-electron microscopy (cryo-EM) structures of the Prime editor in various states, providing a structural framework for understanding this innovative genome engineering system.
Using cryo-EM technology, the researchers analyzed the structures of the Prime editor complex in different states, including pre-initiation, initiation, elongation, and termination. They successfully obtained high-resolution structures of the Prime editor complex in multiple states, revealing its dynamic changes during the guided reverse transcription process. Based on structural information, the researchers designed pegRNA variants and Prime editor variants. They also truncated and fused M-MLV RT, successfully developing a smaller Prime editor variant (PECO-Mini). This variant maintains editing efficiency while improving AAV vector titers and Prime editing efficiency. Activity tests showed that the engineered Prime editor variant has comparable activity to the original Prime editor in vitro. This study reveals the structural characteristics of the Prime editor complex in different working states, providing important information for understanding its molecular mechanism.

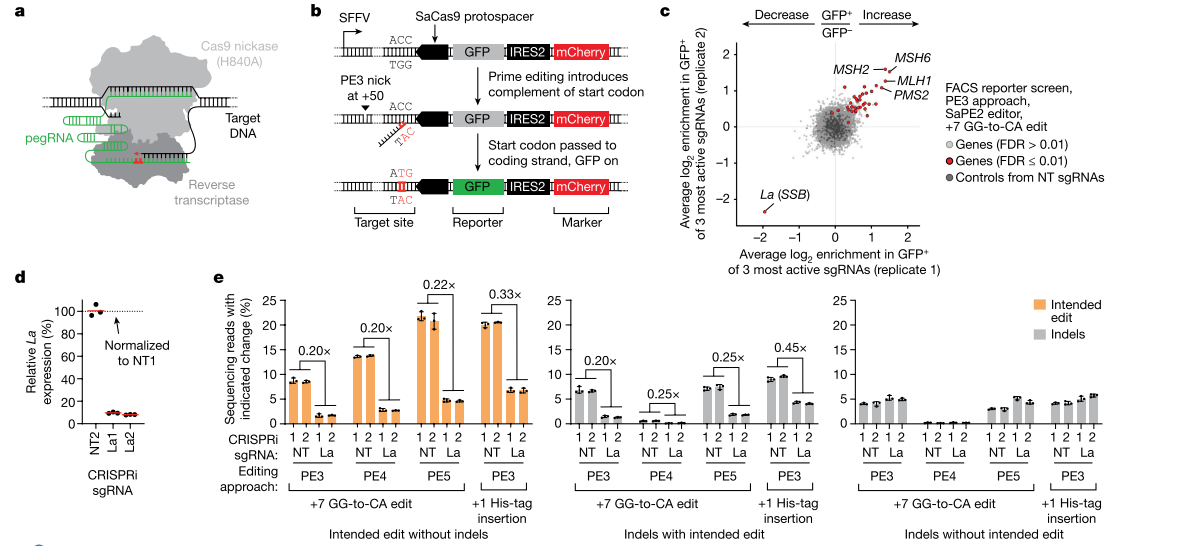
Figure 2: Cryo-EM Structure of Prime Editor
Original Article Link: <https://doi.org/10.1038/s41586-024-07259-6>
While many studies focus on improving the system performance of the Prime editor to enhance its editing efficiency, there is still limited understanding of how Prime editing works within the cellular environment and how its interaction with the cellular context affects editing results. Researchers have attempted to improve Prime editor efficiency by identifying other cellular determinants that influence it and conducting related studies.
Through genome-scale CRISPR interference (CRISPRi) screening, researchers discovered a key Prime editing promoter - the small RNA-binding exonuclease protection factor La. Studies have shown that La protein binds to the 3' polyU sequence of pegRNAs through its N-terminal domain, thereby promoting Prime editing. This effect of La protein is effective for different types of editing (substitution, insertion, deletion) and different cell types. Based on this discovery, the researchers developed a new Prime editor protein (PE7), which fuses the La's RNA-binding N-terminal domain to the PEmax editor. The PE7 editor significantly improves Prime editing efficiency when using expressed pegRNAs, engineered pegRNAs (epegRNAs), and synthetic pegRNAs optimized for La binding.
EDITGENE, leveraging Prime Editing technology, has comprehensively upgraded its Bingo™ platform for prime editing site-specific mutagenesis. Drawing from extensive experience in over a thousand gene editing Contract Research Organization (CRO) projects, the company has distilled, optimized, and enhanced its approach, resulting in significantly higher success rates compared to conventional methods of gene point mutation. EDITGENE now offers precise and efficient gene point mutation cell line construction services to a broad spectrum of scientific research enterprises.
Recent Blogs
2.[Frontier Information] CRISPRi Screening Boosts the Discovery of Key Targets in Tumor Immunity
3.【Cutting-Edge News】New Trends in Gene Editing - FGFR1, BRD4, and BACE1 Knockout Cells
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






