[Literature Review] Exploring the Functional Conservation of Cross-Species lncRNAs Using Cas12a Knockout Technology
In January of this year, researchers from Tsinghua University and Peking University published a study titled "Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human" in Nature Genetics, a journal with an impact factor of 30.8. The study employed CRISPR-Cas12a technology to genetically knockout coPARSE-lncRNAs in human cells that were computationally predicted to have zebrafish orthologs, resulting in defects in cellular proliferation. To assess functional conservation, they performed rescue experiments (KO-rescue) by reintroducing the predicted zebrafish orthologs into cells with the human lncRNA knocked out, attempting to restore the cellular function. Similar experiments were conducted in zebrafish embryos, thereby demonstrating the functional conservation of lncRNAs across species. These findings shed light on the role of lncRNAs in cellular proliferation and development, potentially offering novel therapeutic strategies for cancer treatment.
Long non-coding RNAs (lncRNAs), defined as transcripts longer than 200 nucleotides without protein-coding potential, serve as regulatory elements in numerous physiological processes and are implicated in various diseases. Despite their known importance, the functions of most lncRNAs remain elusive, primarily due to their limited sequence conservation, which may suggest that many lncRNAs lack functional relevance. Comparative sequence analysis is a valuable tool for elucidating lncRNA evolution and function, yet the majority of lncRNAs exhibit minimal sequence similarity, and their evolutionary patterns differ fundamentally from those of protein-coding genes. This necessitates innovative approaches to identify lncRNA orthologs in distantly related species.

I. Homology Assessment of Candidate lncRNA Orthologs and Identification of coPARSE-lncRNA Evolutionary Orthologs
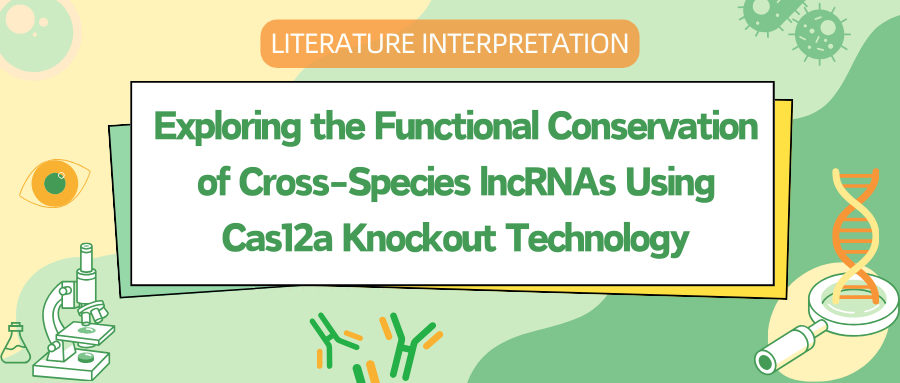
To investigate lncRNA homology, researchers annotated lncRNA datasets from six vertebrates, including bovines, opossums, chickens, lizards, frogs, and zebrafish, complementing the existing high-quality annotations of human and mouse lncRNAs in the GENCODE project. They observed extensive overlap between their annotated lncRNAs and those from five additional public sources. The 20,688 to 42,725 candidate lncRNAs across these six vertebrates consistently displayed lower protein-coding potential, lower expression levels, and higher tissue specificity compared to protein-coding genes, with significantly low sequence conservation. In pairwise BLAST analyses of eight vertebrates (the six mentioned plus humans and mice), only 0.3-3.9% of lncRNAs from one species showed detectable sequence similarity to lncRNAs from another, a much lower rate than for protein-coding genes (40-90%).
Synteny analysis, which identifies genomic regions with shared evolutionary origins, was proposed to aid in the identification of conserved lncRNAs. Researchers thus developed a predictive model based on synthetic recognition of human lncRNA orthologs, using protein-coding ortholog pairs and their associated scores as the training set to predict lncRNA orthology. This analysis revealed thousands of orthologous human lncRNA genes in other species, with the genomic context of the identified synthetic lncRNA candidates closely resembling that of orthologous protein-coding genes.
RNA-binding proteins (RBPs) play a pivotal role in RNA regulation, and researchers postulated that conserved patterns of RBP binding sites could provide valuable insights into identifying functionally conserved lncRNA orthologs. To address this, they established RBP binding motif libraries for the eight species under investigation and predicted 570 to 5,564 human lncRNA orthologs in other vertebrates, designating these as coPARSE-lncRNAs. Among these, 570 human coPARSE-lncRNAs were found to have predicted homology with zebrafish, demonstrating that incorporating conserved RBP binding site data significantly enhances the accuracy in predicting potential lncRNA orthologs.
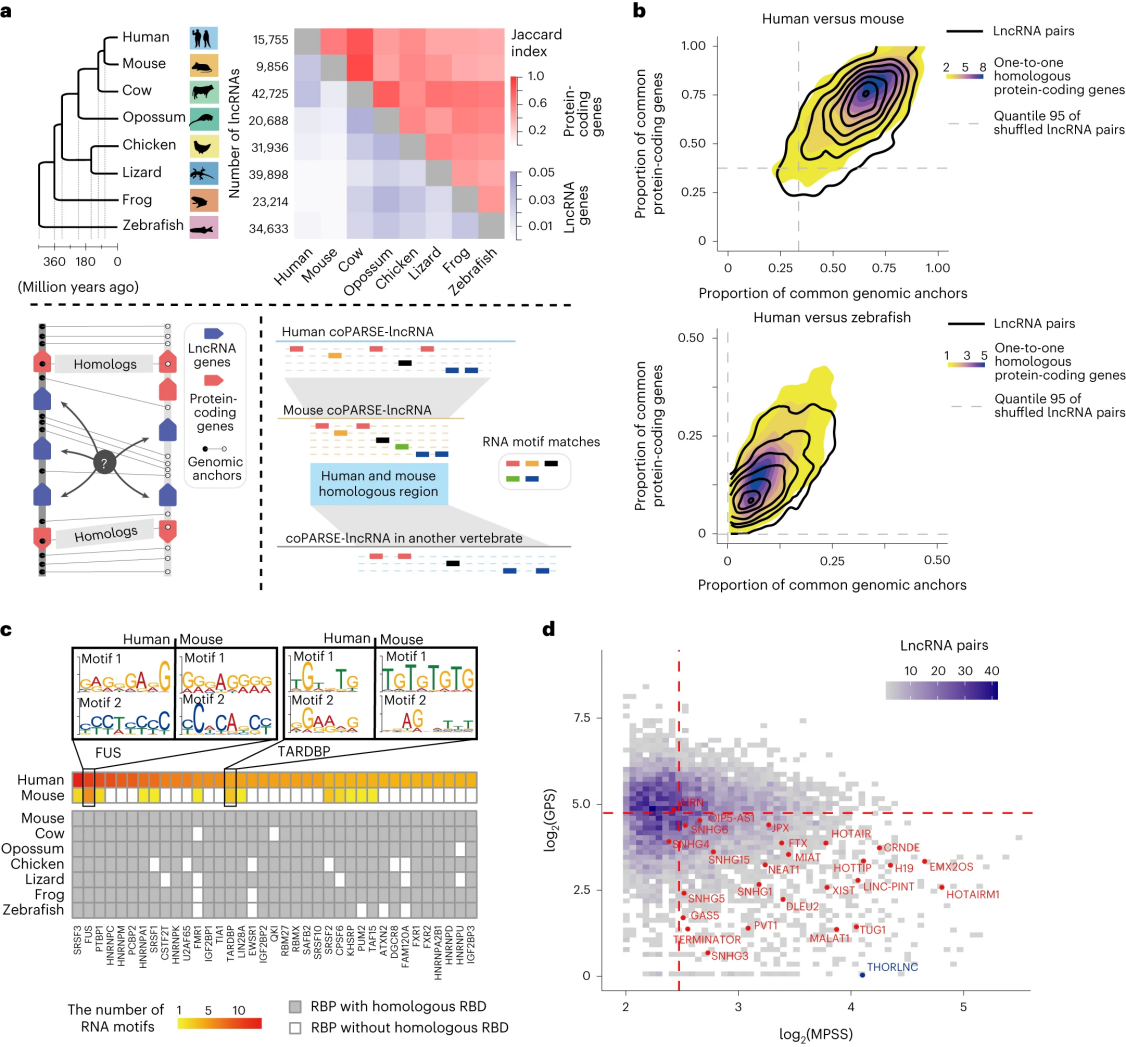
Figure 1: Identification of coPARSE-lncRNAs and Their Homologs in Vertebrates
II. Evolutionary and Functional Characteristics of lncRNA Homologs
Researchers categorized coPARSE-lncRNAs and their homologs into two groups: the homolog_ss group, which consists of 605 pairs of coPARSE-lncRNA homologs exhibiting high sequence similarity (>50%); and the homolog_nss group, which includes an additional 4,959 pairs of coPARSE-lncRNA homologs with low or no sequence similarity. Upon comparing the sequence conservation of coPARSE-lncRNA homolog pairs across both groups, it was discovered that the homolog_ss group demonstrated significantly higher conservation than the homolog_nss group, specifically within the human-mouse and human-zebrafish comparisons. For both the homolog_ss and homolog_nss groups, the density of common single-nucleotide polymorphisms (SNPs) or the frequency of major alternative alleles was notably lower in the base sequence regions compared to non-base sequence regions. These findings suggest that the predicted base sequence regions of coPARSE-lncRNAs have undergone greater selective pressure than non-base sequence regions.
In comparison to random lncRNAs, coPARSE-lncRNA homologs exhibit relatively higher similarity in histone modification patterns and comparable tissue expression profiles across different species, surpassing other random homolog lncRNA pairs. Additionally, through genome-wide association studies, it was observed that 270 out of 570 human coPARSE-lncRNAs are located in disease-associated genomic regions, a proportion higher than other human lncRNAs. Human coPARSE-lncRNAs are enriched with disease-related mutations compared to random lncRNAs, making their expression more susceptible to dysregulation in cancerous tissues.
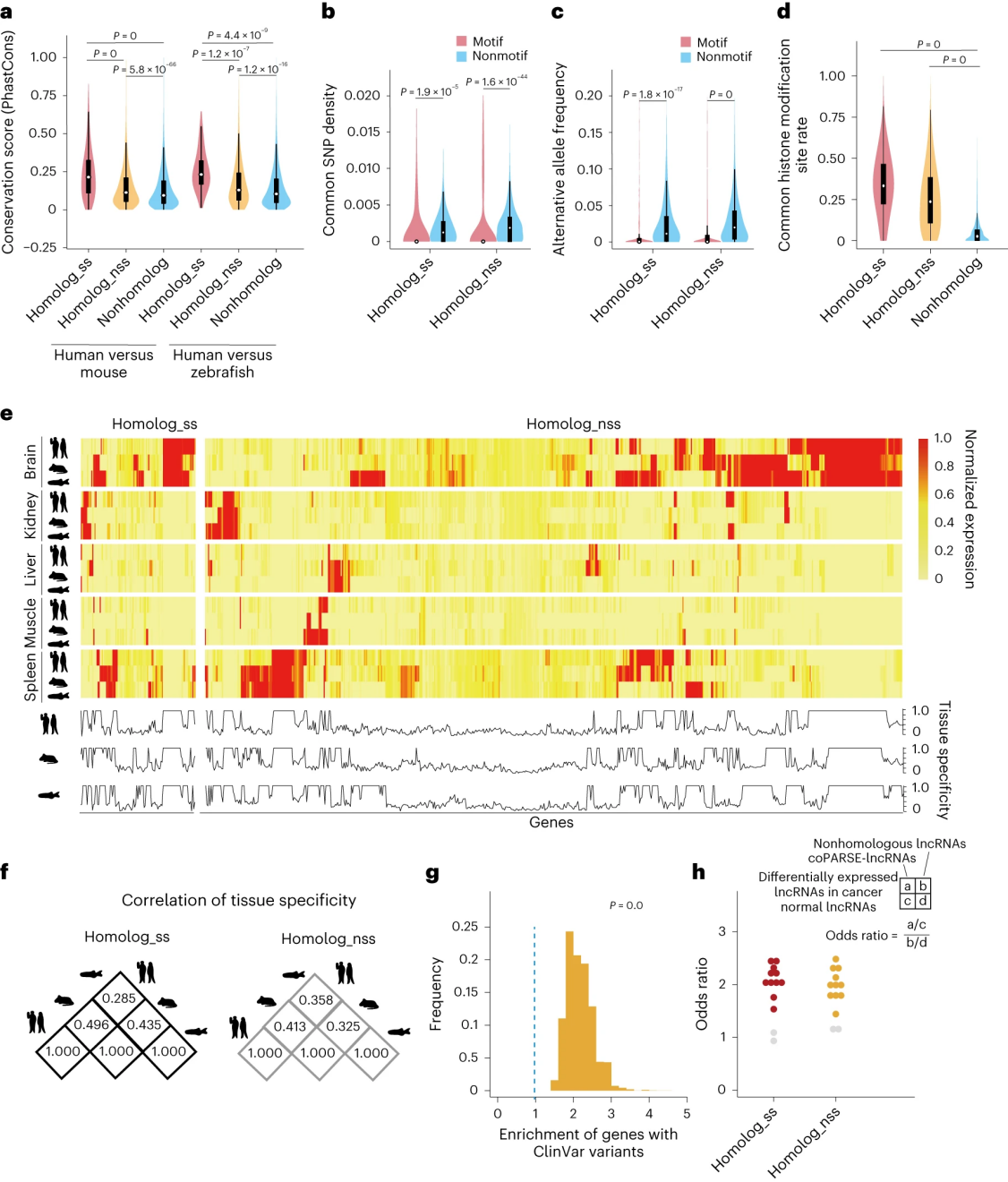
Figure 2: coPARSE-lncRNAs and Their Predicted Homologs Share Similar Evolutionary and Functional Characteristics
III. CRISPR Screening to Identify lncRNAs That Promote Proliferation
To functionally characterize coPARSE-lncRNAs, researchers conducted cell proliferation assays using cancer cell lines. They targeted 574 human lncRNAs that are highly expressed in human cancer samples, including 249 coPARSE-lncRNAs with predicted homologs in zebrafish. For each of the 574 lncRNAs, 20 pairs of crRNAs were designed. The library was introduced into three cancer cell lines (HeLa, Huh7, and MCF7) stably expressing Cas12a via lentiviral transduction. Green fluorescent protein (GFP)-positive cells were selected for propagation to perform a broad CRISPR KO screen.
Across the three cancer cell lines, the researchers identified 167 lncRNAs (including 75 coPARSE-lncRNAs) that showed significantly reduced crRNA abundance on days 15, 30, and 45 compared to day 0. Eighty-two percent of the crRNAs targeting these genes were depleted, with limited overlap between different cell lines. To validate the screening results, several negatively selected coPARSE-lncRNAs were investigated in detail. Compared to a positive control lncRNA, RNY1, and two candidate coPARSE-lncRNAs (RP1-212P9.3 and AL355075.4), the proliferation rate of cells with these lncRNAs knocked out by Cas12a was significantly reduced. shRNA knockdown of three coPARSE-lncRNAs (RP1-212P9.3, AL355075.4, and RP11-563J2.3) confirmed their role in promoting cell proliferation. Knocking out the protein-coding gene OPRD1, which partially overlaps with RP1212P9.3, did not affect cell proliferation. Through CRISPR Cas12a knockout screening, conserved coPARSE-lncRNAs that regulate cancer cell proliferation were identified.
Figure 3: CRISPR-Cas12a Screening and Validation of Functional Conservation in coPARSE-lncRNA
IV. Functional Validation of the Protective Role of lncRNA Homologs
To explore the functional conservation of coPARSE-lncRNA, researchers employed a CRISPR-Cas12a KO-rescue system. After successfully testing doxycycline (Dox)-inducible ectopic gene expression, they transfected cancer cells expressing Cas12a with lentiviral particles targeting 21 human lncRNAs, followed by transfection of rescue sequences from zebrafish homologs into Cas12a-expressing cancer cells. Proliferation assays revealed that, except for RP11-20I23.6, proliferation rates of all selected coPARSE-lncRNA no-Dox groups decreased by 30-70% compared to the control. In cells grown in Dox-containing medium, the proliferation rates of five zebrafish homologs increased, indicating functional compensation for proliferation promotion. For instance, the ectopic expression of the zebrafish homolog region of coPARSE-lncRNA RP1-212P9.3 partially rescued the proliferation defect caused by RP1-212P9.3 KO, while expression of a control fragment of the firefly luciferase gene with matching length did not have a rescuing effect.
The researchers also assessed the potential functional conservation of predicted coPARSE-lncRNA homologs in early zebrafish embryonic development. For four coPARSE-lncRNAs identified in the rescue experiments (RP1-212P9.3, RP11-1055B8.4, RP11-429B14.1, and RP11-223I10.1), they knocked out the predicted homologs in early zebrafish embryos. Observable developmental delays were observed, and the introduction of the sense sequence of the human coPARSE-lncRNA homologs rescued the delay, allowing normal development. These findings suggest a conserved regulatory role for coPARSE-lncRNA across distantly related species.

Figure 4: Functional Validation of coPARSE-lncRNAs
V. Significant Overlap Exists Among the coPARSE-lncRNAs Homologous Interaction Groups
To test whether coPARSE-lncRNAs homolog pairs interact with the same RBPs, researchers conducted RNA pull-down and mass spectrometry (MS) analyses on RP1-212P9.3 and RP11-1055B8.4 to detect interacting proteins with HeLa cell lysate for human, mouse, and zebrafish lncRNA homologs. Principal Component Analysis (PCA) of the pulled-down proteins revealed that, within the lysate, the binding protein profiles of coPARSE-lncRNA homolog pairs are closer to each other compared to different lncRNAs from the same species, strongly supporting the similarity in binding protein spectra among coPARSE-lncRNA homolog pairs. Immunoblotting confirmed that the human coPARSE-lncRNA RP1-212P9.3 and its mouse and zebrafish homologs all interact with CAPRIN1, TARDBP, and NONO.
RNA pull-down experiments identified 6 and 5 RBPs from an RBP library for binding to RP1-212P9.3 and RP11-1055B8.4, respectively, for motif pattern analysis. Among the identified RBPs, 3 out of 6 for RP1-212P9.3 and 2 out of 5 for RP11-1055B8.4 were accurately predicted, and the motifs between homolog pairs matched well.
Researchers also performed RNA pull-down and MS analyses on RP1212P9.3 homologs in mouse cells and early zebrafish embryos. The results showed a strong correlation and extensive overlap in the enriched RBPs of RP1212P9.3 homologs in human HeLa cells, mouse V6.5 cells, and zebrafish embryos. Furthermore, the researchers evaluated the common proteins of RP1-212P9.3 homologs in the corresponding species' cells. Compared to the baseline, there was a relatively high overlap between the pulled-down proteins of RP1-212P9.3 and its homologs. Proteins pulled down by RP1-212P9.3 homologs showed enrichment for functions related to translation and cell proliferation.
Five additional complementary experiments were conducted using predicted human homologous RP1-212P9.3 fragments containing different putative RBP binding sites to rescue the early developmental delay defect in zebrafish caused by TCONS_00107744_zbf knockdown. The results indicated that only the fragment with an intact NONO binding site could rescue the developmental delay in TCONS_00107744_zbf knocked-down zebrafish embryos, demonstrating the specificity of the RBP (NONO) binding site for the rescue fragment. Two KO-rescue experiments were also performed on coPARSE-lncRNAs (RP1-212P9.3 and RP11-1055B8.4) in HeLa cells, revealing that mutations in the NONO binding site and IGF2BP2 binding site abolished the rescue effect of the TCONS_00107744_zbf and TCONS_00075010_zbf fragments, respectively. Combined with the results from the zebrafish rescue experiments, these findings suggest that the interaction of coPARSE-lncRNA homologs with specific RBPs is functionally relevant.
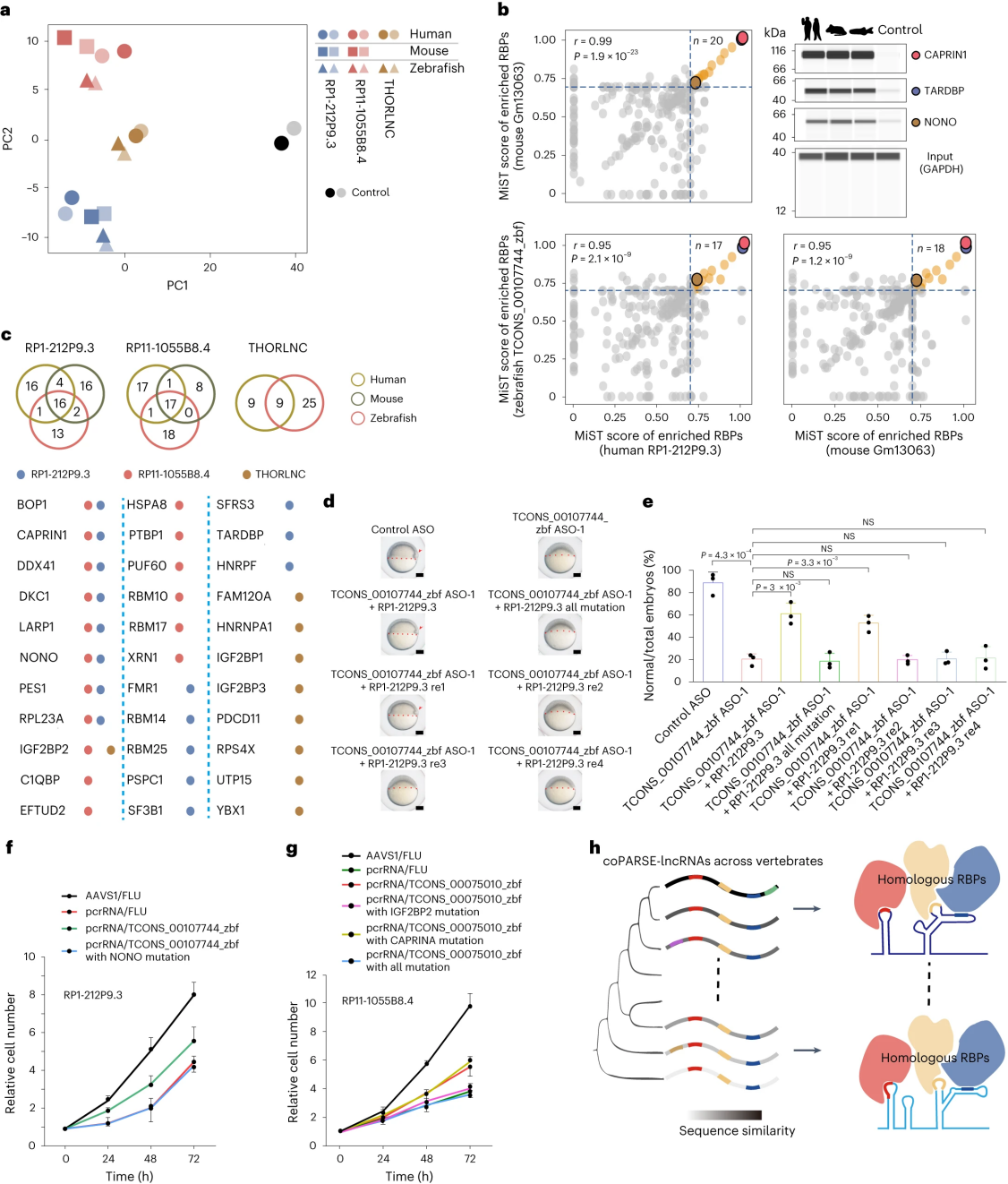
Figure 5: Identification and Functional Analysis of RBP Interaction Groups for Two coPARSE-lncRNAs
In summary, researchers utilized CRISPR Cas12a gene knockout technology and KO-rescue techniques to introduce predicted zebrafish homologs into cells with knocked-out human lncRNAs and inject human homolog lncRNA fragments into embryos with knocked-down zebrafish lncRNA homologs. This successfully repaired the developmental delay caused by the knockout/knockdown, demonstrating the functional conservation of lncRNAs from zebrafish to humans. This provides a new perspective on the evolution of lncRNA functions.
Original article link: https://doi.org/10.1038/s41588-023-01620-7
Recent Blogs
1.[Frontier Information] CRISPRi Screening Boosts the Discovery of Key Targets in Tumor Immunity
2.【Cutting-Edge News】New Trends in Gene Editing - FGFR1, BRD4, and BACE1 Knockout Cells
3. A talk about the Past and Present of Gene Editing Technology: ZFN, TALEN and CRISPR
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com






