[Star of the Month] Ubiquitination-Related Libraries, Lipid Metabolism Libraries, and Mouse Whole Genome Knockout Librari
CRISPR library screening is a high-throughput gene screening method based on CRISPR/Cas9 technology. It involves constructing a library containing thousands of sgRNAs and cloning these sgRNAs into lentiviral vectors to infect target cells at a low MOI. This approach ensures that each cell is infected by only one sgRNA, enabling functional screening of genes corresponding to different sgRNAs. In this article, EDITGENE recommends three of the most popular CRISPR libraries currently available and provides interpretations of relevant research literature on these libraries, hoping to contribute to your scientific research journey.
1. CRISPR-Cas9 Knockout Library Screening to Explore Key Genes Regulating LC3B Ubiquitination
Link to original text: https://doi.org/10.1016/j.jbc.2021.100405
Autophagy is a cellular degradation pathway crucial for maintaining cellular homeostasis, and its defects are associated with the pathogenesis of various diseases, including neurodegenerative diseases, myocardial lesions, cancer, type 2 diabetes, and immune system diseases. LC3B undergoes ubiquitination catalyzed by ubiquitin-activating enzymes and ubiquitin-conjugating enzymes, leading to its degradation through the proteasome pathway and thus reducing LC3B levels and autophagy activity under stress conditions. However, the mechanisms that counteract this process remain unclear.
Researchers used a CRISPR ubiquitination knockout library to screen 661 genes related to ubiquitination, aiming to identify genes that regulate the levels of the autophagy protein LC3B. The results showed that knockout of the deubiquitinating enzyme USP10 (Ubiquitin Specific Peptidase 10) gene reduced intracellular protein levels of both LC3B-I and LC3B-II, leading to increased degradation of LC3B through the proteasome pathway. USP10 can deubiquitinate LC3B, thereby increasing LC3B levels and autophagy activity. The identification of USP10 as a new LC3B regulator through CRISPR ubiquitination knockout library screening reveals its crucial role in regu.
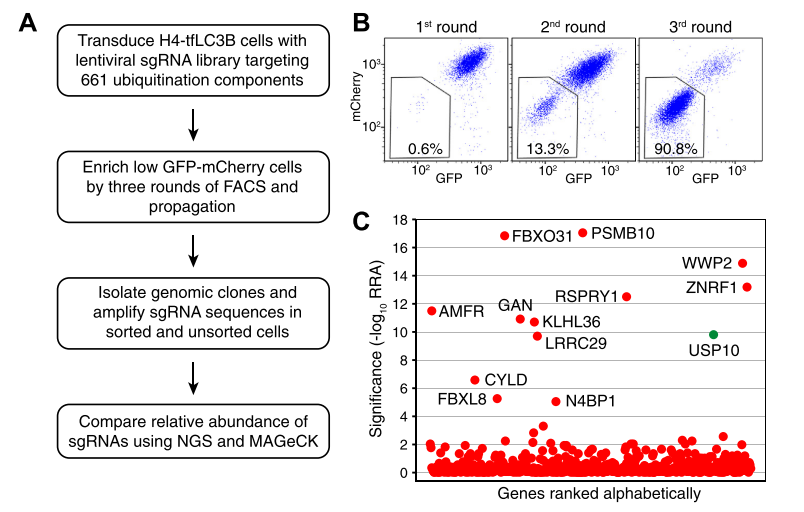
Figure 1: CRISPR-Cas9 KO Screening for Ubiquitination-Related Proteins that Regulate LC3B Levels
Researchers used a custom screening library, Human Lipid Droplet and Metabolism sgRNA Library, to screen genes related to lipid droplets and metabolism, identifying genetic modifiers affecting PLIN2 expression and post-translational stability. They discovered that the E3 ubiquitin ligase MARCH 6 is a factor regulating triacylglycerol (TAG) biosynthesis and lipid droplets by affecting PLIN2 stability. The new genes and regulatory mechanisms identified through human lipid metabolism library screening may become potential targets for treating diseases related to lipid metabolism disorders. By identifying genes that regulate PLIN2 and lipid droplets, the study has enhanced our understanding of how lipid droplets form, degrade and interact with other organelles within cells.
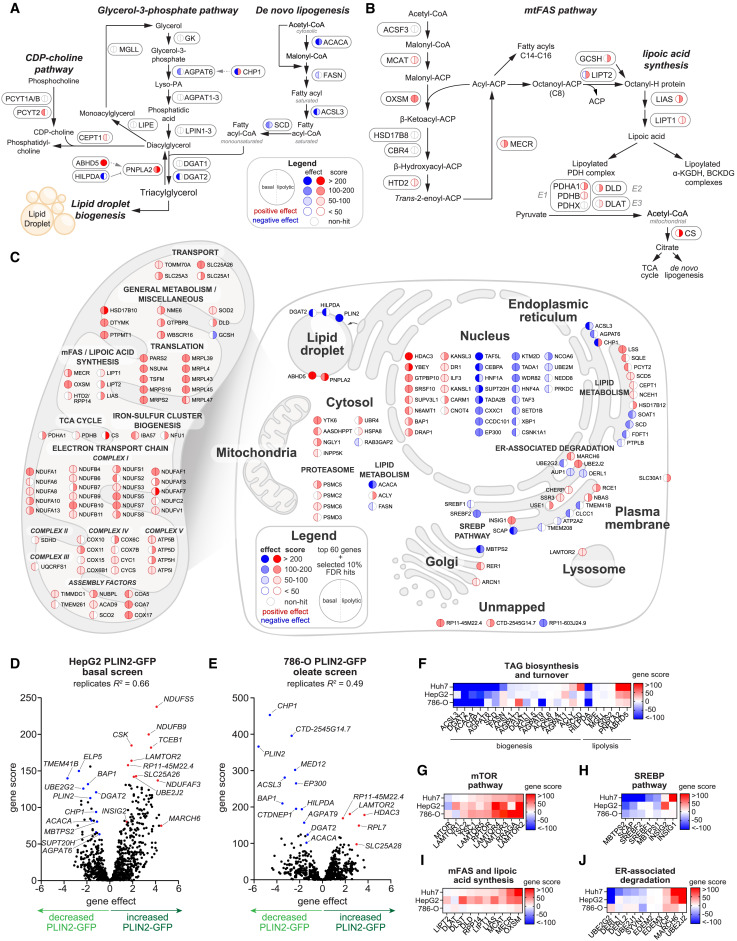
Figure 2: CRISPR-Cas9 Screening for Cell Type and Metabolic State-Dependent Regulators of PLIN2-GFP
3. Genome-wide Knockout Library Screening in Mice Reveals PPP6C as a Positive Regulator of TNF-Induced Necroptosis
Original Link: https://doi.org/10.1038/s41419-022-05076-1
Necroptosis is a form of programmed cell death mediated by specific signaling pathways, often triggered by stimulatory factors such as tumor necrosis factor (TNF) and mediated through the receptor-interacting protein kinase 1 (RIPK1)-RIPK3-mixed lineage kinase domain-like protein (MLKL) signaling pathway. RIPK1, RIPK3, and MLKL, as key effector molecules of necroptosis, have been extensively studied for their roles in regulating necroptosis. However, it remains unclear whether phosphatases are involved in the regulation of necroptosis.
Researchers conducted a genome-wide CRISPR/Cas9 screening using the mouse GeCKOv2 CRISPR knockout pooled library to identify genes that regulate tumor necrosis factor (TNF)-induced necroptosis. PPP6C (the catalytic subunit of protein phosphatase 6) was identified as a positive regulator of TNF-induced necroptosis. The absence of PPP6C protected cells from TNF-induced necroptosis, and this protective effect was dependent on its phosphatase activity. The deletion of PPP6C led to hyperactivation of TAK1 and reduced RIPK1 kinase activity. In the gastrointestinal tract of mice, heterozygous deletion of PPP6C alleviated necroptosis-related tissue damage and inflammation. The mouse GeCKOv2 CRISPR knockout library elucidated the molecular mechanism of PPP6C regulating RIPK1 kinase activity through the TAK1-IKK signaling axis, providing a theoretical basis for the development of small-molecule drugs targeting PPP6C or its regulatory pathways.
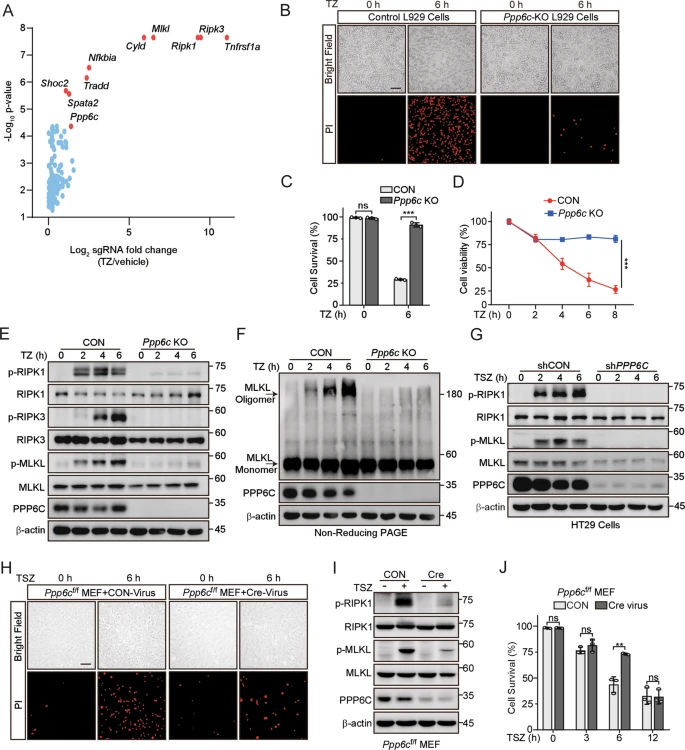
Figure 3: Deletion of PPP6C Prevents TNF-Induced Necroptosis
EDITGENE is proud to have the most comprehensive CRISPR plasmid library, offering knockout, activation, and inhibition options. Our current promotional offer is in full swing, with high-quality library plasmids. These plasmids have a coverage rate of ≥99% and a uniformity of <10! CRISPR Plasmid Library Promotional Details >>>
You may be interested in:
【New Trends in Gene Editing】CTLA4, MUC1, and mTOR Knockout Cells
Pre-experiment essentials: A walkthrough of the CRISPR Assay
[Frontier Information] The first AI gene editor has successfully edited the human genome
Follow us on social media

