The Knits and Grits behind single-cell CRISPR screening
I. Traditional CRISPR library screening technology
Since the emergence of CRISPR library screening technology, it has become the best solution for large-scale gene screening to establish the relationship between genes and phenotypes in a high-throughput manner. In the traditional CRISPR screening technology, sgRNA (single guide RNA, sgRNA) targeting different genes is synthesized according to the research purpose, and lentivirus is used as a vector to transduce the gene editing system into the screened cells to obtain a mixed library of cells with different editing events. The cells were screened, cultured, and enriched for specific phenotypes. The sgRNA sequences in the enriched cells can be amplified to screen for genes associated with the phenotype. Conventional CRISPR screening techniques have been widely used for high-throughput target gene screening and targeted drug discovery.
II. The principle and advantages of single-cell CRISPR screening
Traditional CRISPR screening only tells us that the target gene obtained from the process is related only to the phenotype and not through the gene's regulatory network. Single-cell CRISPR screening is a good solution when we want to screen the target genes that produce the phenotype and also want to know the effect of each target gene on other signaling pathways.
On the other hand, single-cell CRISPR screening is based on its single-cell transcriptome sequencing technology. Through single-cell sequencing of CRISPR screening libraries, a single cell's sgRNA and transcriptome data information is obtained. Then, the correspondence between the phenotype and the relevant affected genes is determined through the sgRNA indexes in the libraries. Based on such sequencing ideas, various sequencing technologies have been developed since—for example, Perturb-Seq, CRISP-Seq, CROP-Seq (CRISPR droplet sequencing), and Direct capture PerturbSeq.
These technologies enable detection at single-cell resolution and directly establish relationships between sgRNAs and other gene expression profiles and phenotypes. Compared to traditional CRISPR screening techniques or individual gene editing cell models, they have higher validation efficiency. Below are descriptions of these classic single-cell CRISPR screens.
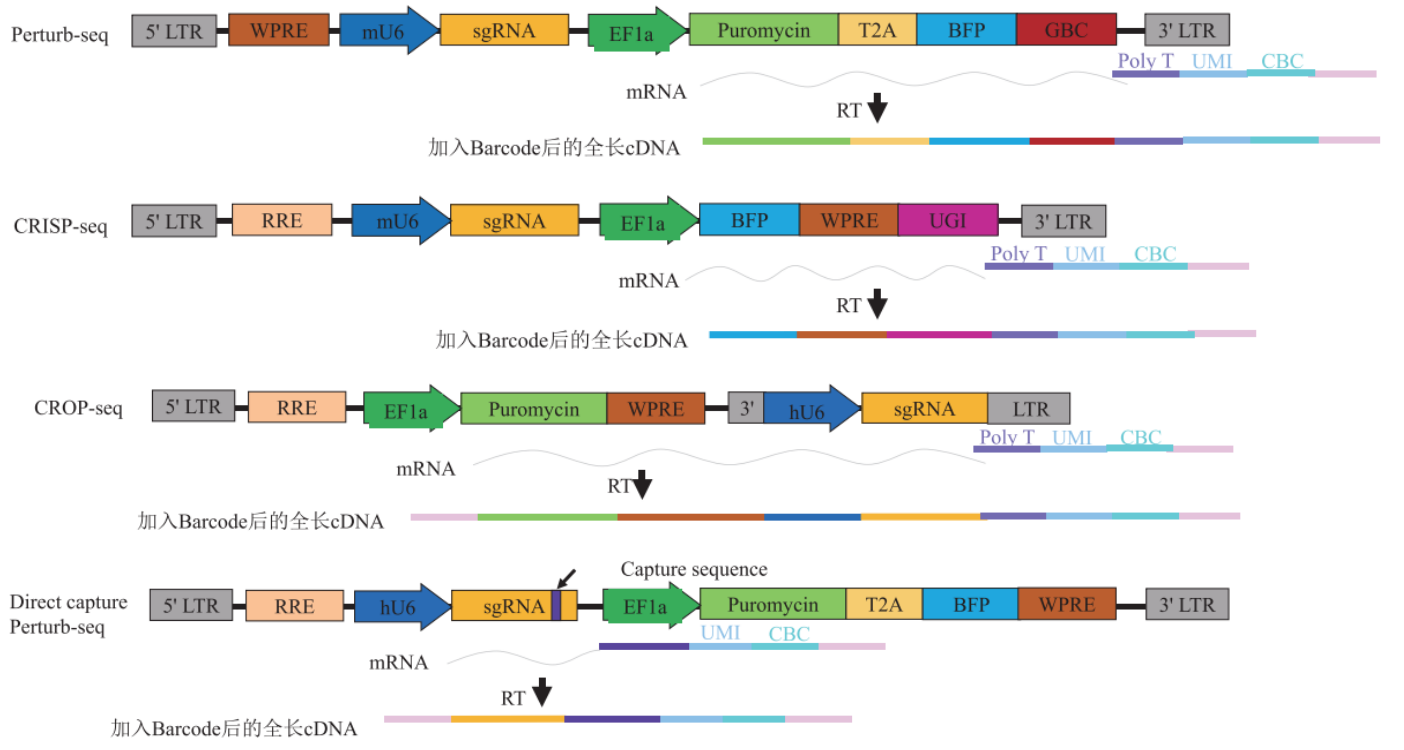
III. Single-cell CRISPR screening methods
1. Perturb-Seq
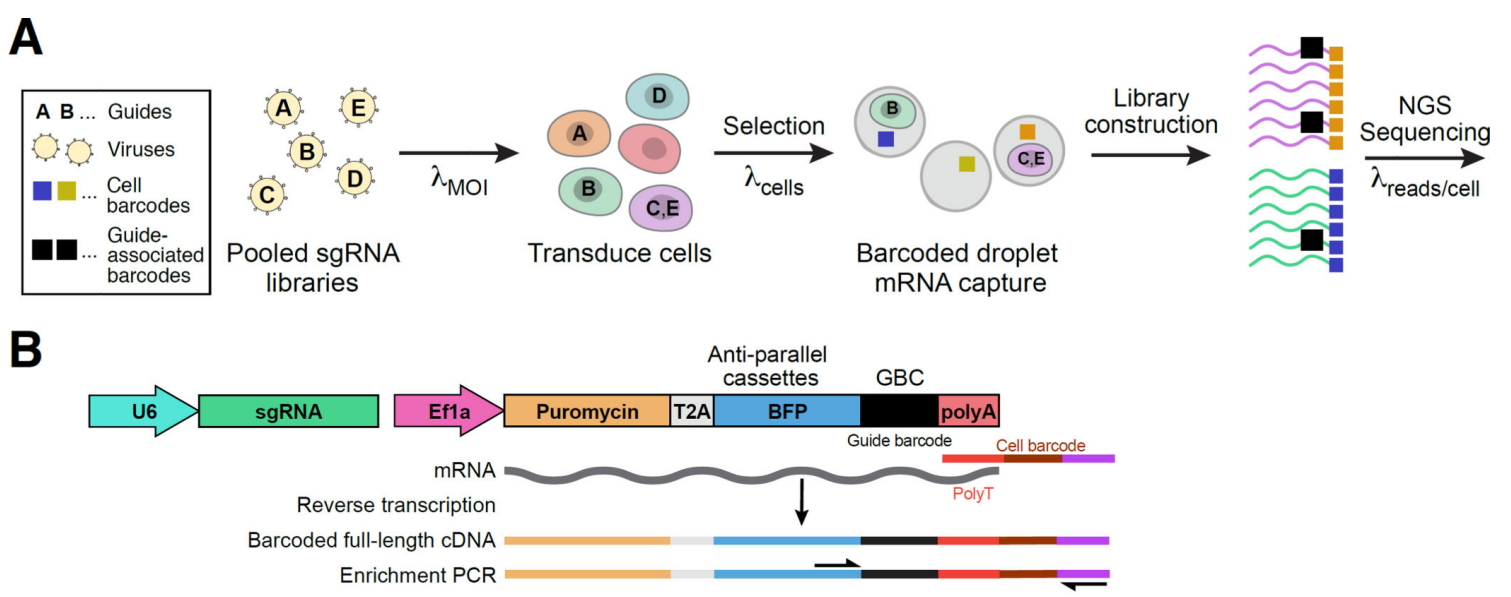
Combined with the high-throughput genomic function analysis method of single-cell sequencing, Perturb-Seq is an enhanced and optimized screening method from the traditional CRISPR screening. [2]. The method designs a guide barcode (GBC) for each sgRNA and adds an expression cassette for the GBC to the CRISPR lentiviral vector. Since the expression cassette can be used in a single-cell sequencing system, the data obtained contains the GBC and the transcriptome. From this, the relationship between perturbed genes, regulatory networks, and expression is analyzed for each cell. In addition, the expression cassette contains blue fluorescent protein (BFP) and puromycin resistance genes for use in flow cytometry sorting or antibiotic selection of transduced cells.
2. CRISP-Seq
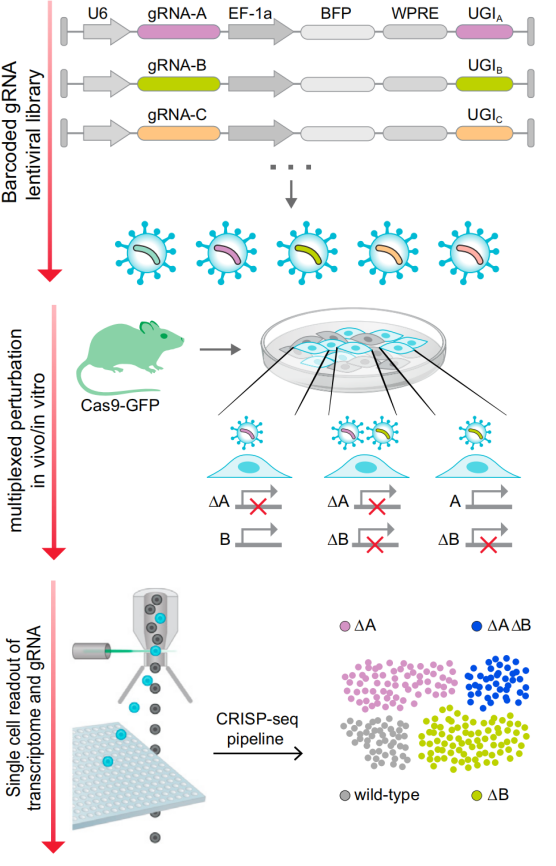
The principle of action of CRISP-Seq is similar to that of Perturb-Seq. It is an expression cassette in CRISPR lentiviral vectors with a polyadenosine unique guide index (UGI), which acts similarly to GBC to label sgRNAs. Combining the UGI with single-cell transcriptome data, single-cell sequencing enables in-depth and comprehensive phenotypic analysis of gene perturbations and the study of their functions and interactions in a single experiment [3].
3. CROP-Seq
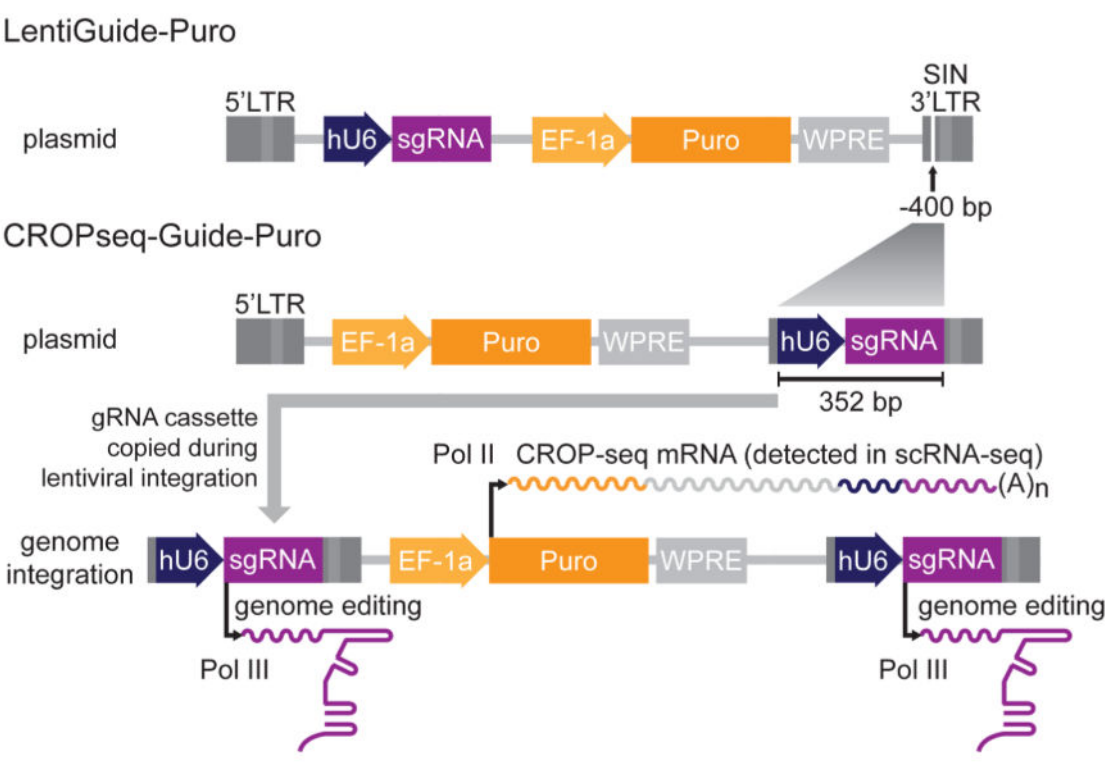
The lentiviral vector structure of CROP-Seq is an insertion of a U6-gRNA expression cassette in the 3′ LTR region after the Puro resistance gene. RNA polymerase III transcribes the U6-gRNA with this design and performs the gene editing function. While RNA polymerase II transcribes the puro resistance gene, puromycin mRNA will contain the sequence of the U6-gRNA expression cassette. In the subsequent single-cell sequencing, the information of sgRNA can be obtained directly, which significantly simplifies the subsequent operation [4]. In addition, during lentiviral reverse transcription and integration, the entire U6-gRNA expression cassette is replicated and inserted into the 5′ LTR but does not interfere with the second copy upstream of the EF-1a promoter.
4. Direct capture Perturb-Seq
Perturb-Seq, CRISP-Seq, and CROP-Seq have been able to analyze the relationship between target genes, regulatory networks, and phenotypes on a single-cell basis, but these methods still have drawbacks:
(1) In Perturb-Seq and CRISP-seq, GBC or UGI labeling of sgRNA is required to detect the corresponding sgRNA, but errors in sequencing, raw-letter analysis, etc., and a certain probability of error in pairing GBC or UGI with sgRNA exist.
(2) The one-to-one correspondence between GBC or UGI and sgRNA means that both the marker sequence and the sgRNA must be cloned into lentiviral vectors, greatly increasing the difficulty of producing sgRNA libraries.
(3) CROP-Seq involves adding a large fragment of the U6 sgRNA expression cassette to the 3’LTR region after the Puro resistance gene, which limits the addition of multiple sgRNAs.
Direct Capture PerturbSeq, on the other hand, solves these problems well. Direct Capture PerturbSeq is based on 10×Gebomics' 5’-end and 3’-end library construction method, and two detection schemes for sgRNAs at the 5’-end and 3’-end have been developed [5]. In the 5’-end detection scheme, reverse transcription primers are designed for the constant sequence behind the sgRNA, cDNA of the sgRNA sequence is synthesized by reverse transcription, and the sgRNA cDNA, together with other mRNA cDNAs, is bound to the TSO sequences on the gel beads, which are ligated to the cell barcode (CBC) and the unique molecular identifier (UMI) of the single-cell transcript, and library sequencing was performed. At the 3′ end of the assay protocol, gel microbeads with CBC, UMI, and reverse transcription primers designed for specific sequences in the constant region, cDNAs of sgRNA sequences were synthesized during reverse transcription, and TSO sequences were added. Libraries were then constructed from the process and sequenced along with other mRNA-cDNAs.
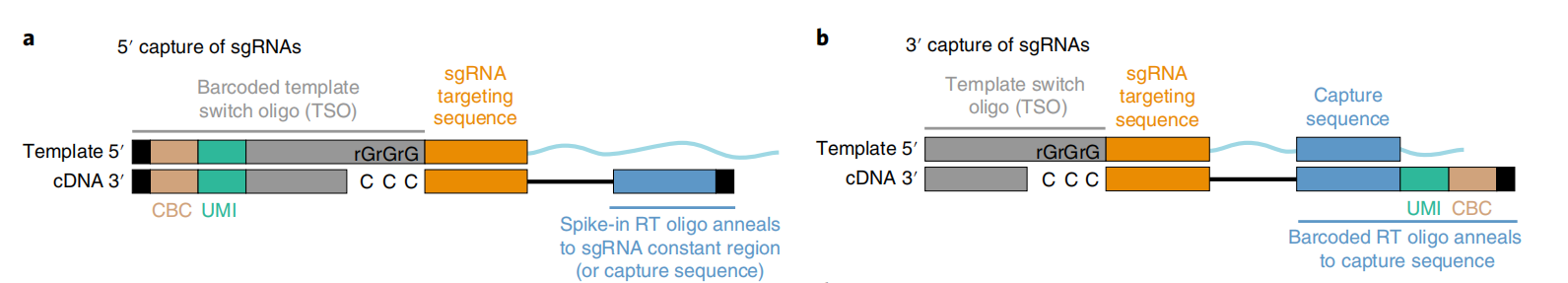
It should be noted that the constant sequence behind the sgRNA, which refers to the sgRNA scaffold, is the secondary RNA structural sequence that anchors the sgRNA to the Cas9 protein. A specific sequence in the constant region is a specific sequence added only to one side of the selected constant sequence without affecting its function and editing efficiency. This sequence is used to design reverse transcription primers.
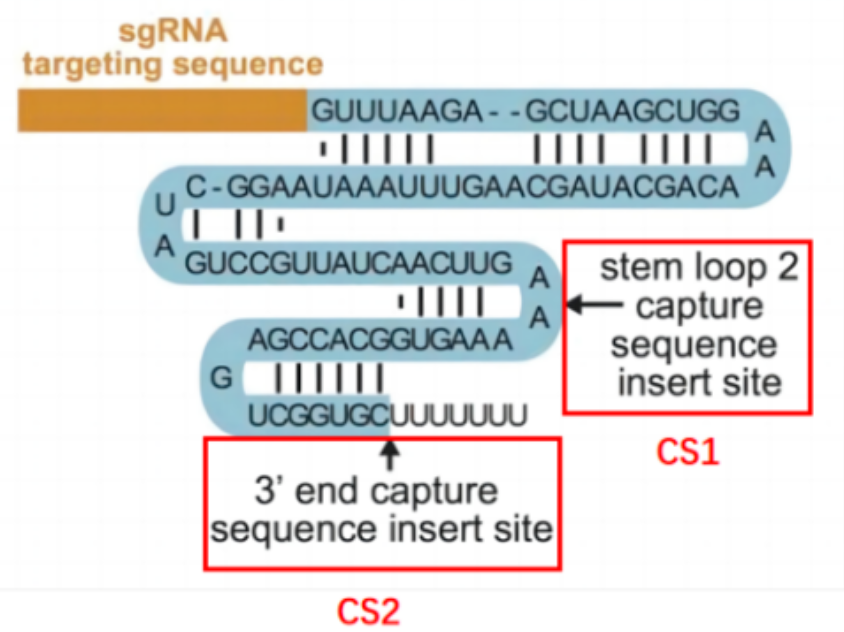
The two library’s construction methods of Direct Capture PerturbSeq are as effective as previous methods such as Perturb-Seq, CRISP-Seq, and CROP-Seq. One lentiviral vector can be loaded with multiple U6-sgRNAs, significantly improving the efficiency of single-cell perturbation experiments.
Summary
CRISPR library screening technology is characterized by high throughput and precise screening of target genes, significantly simplifying the experimental process and saving both time and cost in our research. Combined with single-cell sequencing, the relationship between sgRNA and other gene expression profiles and phenotypes can be directly established based on single-cell high-resolution, making research results more accurate and comprehensive. With continuous iteration and advancement of technology, CRISPR library screening combined with single-cell sequencing are sure to make great contributions to exploring basic life science and treating human diseases.
Editgene has specialized in using CRISPR technology for many years, providing professional CRISPR library screening services (link to the library screening service page) and single-cell sequencing services. The expert team is ready to connect and accelerate your gene function research!
Editgene Newsroom
[MAR Bestselling] CRBN and Myc knockout cell lines; CRISPR whole genome library
[Research Frontier] New trends in CRISPR-- Kras, NLRP3 and BRAF knockout cell lines
[Advanced Materials] Cas13 provides a new strategy for immunotherapy of tendon injuries
References
[1]Dixit A, Parnas O, Li B, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167(7):1853-1866.e17.
[2]Jaitin DA, Weiner A, Yofe I, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167(7):1883-1896.e15.
[3]Datlinger P, Rendeiro AF, Schmidl C, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14(3):297-301.
[4]Replogle JM, Norman TM, Xu A, et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat Biotechnol. 2020;38(8):954-961.

