[Research frontier] CRISPR Detection Trends
The nucleic acid testing for quick specificity detection of infectious pathogens is widely recognized, such as Polymerase Chain Reaction (PCR). However it is expensive due to the expensive equipment and the highly trained Researchers.In this way, CRISPR-Cas system have recently emerged as an alternative to PCR-based diagnosis. CRISPR diagnosis is also known as next-generation molecular diagnostic technology, which mainly utilizes RNA to detect biomarkers in biological fluids(such as blood or urine). By processing the sample preparation (including pre-amplification), target recognition, signal release and testing, the CRISPR/Cas system become the potential diagnostic tool for disease and pathogen detection. It is reported that CRISPR/Cas biosensing systems have the advantages of high resolution, high sensitivity, low cost, simple operation and storage.
In the following, we select and share the latest cases of CRISPR detection applications in humans, animals, and plants to provide some ideas and help for CRISPR detection research.
Colorimetric Detection of Pseudomonas aeruginosa
Pseudomonas aeruginosa(P. aeruginosa), a Gram-negative pathogen, has attracted increasing attention in post-operative infections and poses a new threat to human health. The P. aeruginosa is difficult to be controlled becasue of the wide range of habitats(such as water, air, animals, and humans). Developing a reliable detecion methods for P. aeruginosa is particularly important but still a huge challenge. P. aeruginosa causes long-term chronic diseases by causing bone and joint infections in patients with compromised immune function. In this study, assisted by target identification, the self-primers initiated the production of single-stranded DNA, which can be recognized by CRISPR-Cas12a. Therefore, the trans-cleavage activity of Cas12a enzyme was activated, digesting the silver ion-chelating aptamer sequence. Due to the strong trans-cleavage activity of Cas12a enzyme, the DNA aptamer-based silver ions detection exhibits higher sensitivity. Besides, the limit of detection(LOD) in this method is 21 cfu/mL, which enables a broad application in early diagnosis of infection.
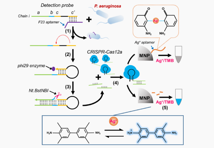
Schematic diagram of colorimetric method based on CRISPR-Cas12a
Reference: Hu, Jiangchun & Liang, Ling & He, Mingfang & Lu, Yongping. (2023). Sensitive and Direct Analysis of Pseudomonas aeruginosa through Self-Primer-Assisted Chain Extension and CRISPR-Cas12a-Based Color Reaction. ACS Omega. 8. 10.1021/acsomega.3c04180.
- EDITGENE provides all the CRISPR raw materials used in the research, such as Cas12a, Cas12b, Cas13a, RPA, reporter. Click to view the list of CRISPR detection raw materials
Developing a Rapid Detection Method for Plant Pathogens
Fusarium head blight (FHB) is a global cereal disease caused by species of Fusarium. Both Fusarium graminearum and Fusarium asiaticum are the causal agents of FHB in China. Asian grass is a dominant species in the middle and lower reaches of the Yangtze River (MLRYR) and southwest China. Therefore, timely detection of Fusarium asiaticum is crucial to controlling the disease and preventing mycotoxins from entering the food chain. This study optimized reaction conditions based on CRISPR Cas12a technology and developed a rapid, sensitive, and cost-effective detection method for Fusarium asiaticum. The optimized method showed specificity in detecting Fusarium asiaticum without detecting any other 14 strains of Fusarium and 3 non-Fusarium species. The limit of detection of this method is 20 ag/µL, and even Fusarium asiaticum in corn and wheat grains can be diagnosed after 3-day inoculation. This method truly provide an efficient and robust detection platform for accelerating the detection.
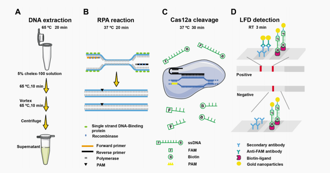
The detection process of the developed RPA-Cas12a-LFD detection system
Reference: Zhang, J.; Liang, X.; Zhang, H.; Ishfaq, S.; Xi, K.; Zhou, X.; Yang, X.; Guo, W. Rapid and Sensitive Detection of Toxigenic Fusarium asiaticum Integrating Recombinase Polymerase Amplification, CRISPR/Cas12a, and Lateral Flow Techniques. Int. J. Mol. Sci. 2023, 24, 14134. https://doi.org/10.3390/ijms241814134
- If you also want to utilize CRISPR detection to diagnosis pathogens, EDITGENE offers a complete solution,which is with high sensitivity, quick detection and strong specificity. For more information, please visit us.
Detection and Serotyping Identification of Streptococcus suis Serotype 2
Streptococcus suis Serotype 2 is an economically important zoonotic pathogen that can cause sepsis, arthritis, and meningitis in pigs and humans. Streptococcus suis Serotype 2 may cause significant economic losses to the pig industry ,threatening the public health. Therefore, developing an accurate and rapid detection method is significant for epidemic prevention and control.
This study developed a detection and serotyping platform named Cards-SSJ/K for serotype 2 of swine streptococcus based on recombinant polymerase amplification technology (RPA) and CRISPR-Cas12a system. The detection method has a detection limit of 10 CFU and a detection time of less than 60 minutes. It has no cross-reactivition with other Serotypes of Swine Streptococcus, closely related Streptococcus or other common pig pathogens. and Cards-SSK can distinguish between serotype 2 and serotype 1/2. The results of Cards-SSJ and qPCR detection of Swine Streptococcus serotype 2 are the same. The results showed that although the reagent cost of Cards-SSJ is relatively high compared to PCR and qPCR, it takes less time and has lower requirements for equipment and personnel. Therefore, it is an excellent method for POCT detection of Swine Streptococcus serotype 2.
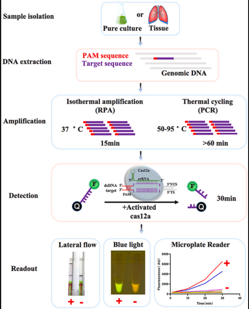
The process of detecting serotype 2 of Swine Streptococcus based on CRISPR Cas12a technology
Refernce: Wang, Lu & Sun, Jing & Zhao, Jiyu & Bai, JieYu & Zhang, Yueling & zhu, Yao & Zhang, Wanjiang & Wang, Chunlai & Langford, Paul & Liu, Siguo & li, Gang. (2023). A CRISPR-Cas12a-based platform facilitates the detection and serotyping of Streptococcus suis serotype 2. Talanta. 267. 125202. 10.1016/j.talanta.2023.125202.
- EDITGENE is specialized on developing and optimizing CRISPR/Cas-based genome editing technologies. The technical team has obtained Cas12 and Cas13 genome editing protein with better-performance through the self-developed Fish-bait purification process,and developed a unique crRNA design system. This has led to the establishment of the FASST detection technology, which greatly improves the cleavage activity of Cas enzyme and CRISPR detection sensitivity.
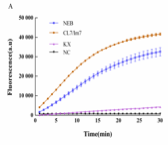
Test of LbCas12a Activity
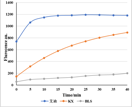
Test of LwaCas13a Activity
Recent Posts:
【OCT Best Seller】XPO5, LRP6, RNF123 Knockout Cell Lines
CRISPR Library Screening Applications

