CRISPR Library Screening Applications
Applications of CRISPR Library Screening
In the past few years, CRISPR technology has become a new gem, and CRISPR library screening is one of its applications. By using the whole genome knockout libraries or sub-pool knockout libraries, sgRNAs of genes related to certain phenotypes will be enriched. This high-throughput functional screening technique has gradually replaced RNAi and cDNA libraries, providing a high-throughput and efficient screening tool for the study of functional genes.
Common CRISPR sgRNA library screening applications are as follows:
· Exploring the mechanism of drug action, identifying and validating drug targets.
· Discovering cancer treatment targets by analyzing upstream and downstream regulatory mechanisms.
· Exploring the therapeutic mechanisms of metabolic diseases by analyzing metabolic pathway regulation mechanisms.
The following 3 cases are typical applications of CRISPR library screening.
Case 1: Screening for new therapeutic targets for hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. The high mortality rate of HCC is due to a lack of biomarkers suitable for early detection, insufficient understanding of HCC heterogeneity, and treatment resistance. Cancer stem cells (CSCs) in HCC are believed to be responsible for initiating heterogeneous tumor lesions, leading to tumor recurrence, metastasis, and treatment resistance. At present, remarkable progress has been made in the identification of liver CSCs based on surface and cellular diagnostic markers. However, CSCs can also be recognized in cell populations that do not express these markers. Therefore, understanding the detailed regulatory mechanisms of the appearance and expansion of CSCs provides an opportunity to improve the prognosis of HCC patients.
By screening the metabolic gene CRISPR knockout library in tumor spheres from HCC cells, it was found that removing SCARB2 inhibited the CSC-like characteristics of HCC cells. In the HCC mouse model driven by myc and induced by DEN (diethylnitrosamine), knockout Scarb2 in liver cells can weaken the initiation and progression of HCC. The binding of SCARB2 and MYC promotes the acetylation of MYC by interfering with the deacetylation of lysine 148 mediated by hdca3, thereby enhancing the transcriptional activity of MYC. Screening of drug databases approved by the FDA (Food and Drug Administration) showed that polymyxin B exhibits high affinity for SCARB2 protein, disrupting SCARB2-MYC interactions, reducing MYC activity, and reducing tumor burden. Our study identified SCARB2 as a functional driver of HCC, indicating that polymyxin B therapy can be an optional treatment for HCC.
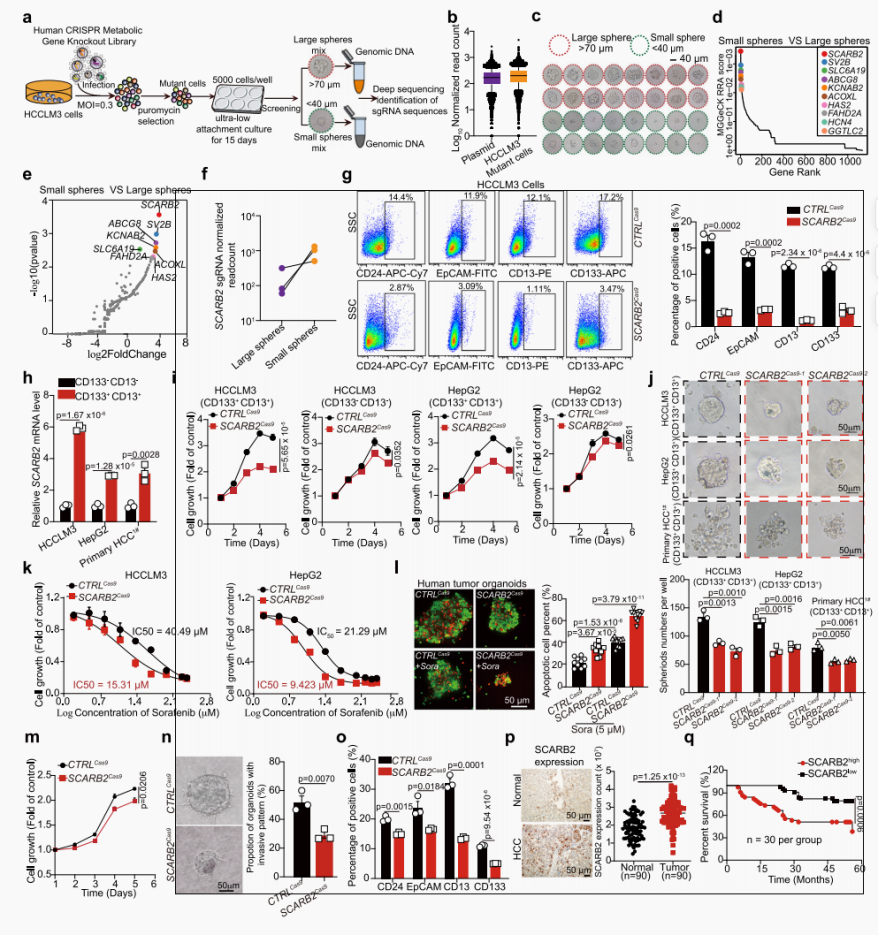
Figure 1. SCARB2 may perform critical pathogenic functions by supporting stem cell-like characteristics in HCC cells
- EDITGENE provides the aforementioned metabolic gene knockout library, lentiviral library only 1050 USD! Check out our pre-made lentivirus libraries>>
- Got a list of genes after CRISPR screening? Why not do as this author did? Generate KO cell lines to study the genes futher! Now KO cell line buy 3 get 1 FREE, knockout whatever gene you want! Visit us for more details>>
Case 2: Library screening helps to understand immune escape in cancer treatment
Immune checkpoint inhibitor therapy has revolutionized cancer treatment, but some cancers, such as acute myeloid leukemia (AML), do not respond or develop resistance. A potential immune escape mechanism involves abnormal presentation of MHC-I class molecular antigens in the main histocompatibility complex, which affects the number and function of killer CD8 T cells, thereby achieving immune escape.
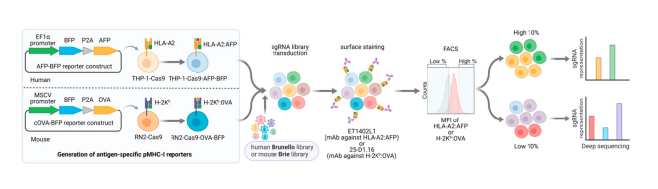
Figure 2. CRISPR screening of tumor antigen presentation related targets
Acute myeloid leukemia (AML) cells originate from myeloid cells that can differentiate into antigen presenting cells, and the antigen presenting mechanisms found on these cell lines are likely to exist in other tumor types. Researchers used the peptide MHC-I guided CRISPR library to screen AML cells. During the screening process, they found that the top ranked negative regulatory factors included surface protein SUSD6, TMEM127, and WWP2. Among them, TMEM127 is a four-pass transmembrane protein that is associated with susceptibility to neuroendocrine tumors and is also considered a Nedd4 family E3 ligase adapter. It is downregulated by the Salmonella effector protein SteD to cleave MHC II. SUSD6 is a single-pass transmembrane protein with no reported immune related function.
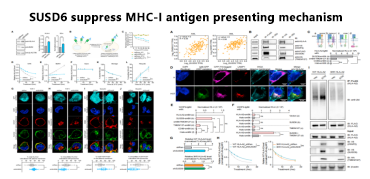
SUSD6 and TMEM127 can directly interact with MHC-I simultaneously and recruit E3 ubiquitin ligase WWP2 to form a quaternary complex. WWP2 mediates MHC-I ubiquitination and lysosomal degradation in the presence of SUSD6 and TMEM127, thereby reducing the expression of MHC-I on the cell surface.
Combining the characteristics of the SUSD6/TMM127/WP2 gene, which is negatively correlated with cancer survival rate, it was found that these key genes regulating MHC-I molecules are potential therapeutic targets for leukemia and solid cancer, and have great value for clinical applications.
- No pre-made library for your study? No worries. EDITGENE provides affordable custom CRISPR screening services. Please visit us for more details>>
Case 3: Library screening helps to discover new ideas for leukemia treatment
Differentiation therapy has been recognized as an effective treatment strategy for acute myeloid leukemia (AML). Although FOXO transcription factors are a promising drug target for differentiation therapy, the efficacy of FOXO inhibitors in vivo is limited. Here, the author found that the pharmacological inhibition of a common cis regulatory element among members of the (FOXO) family successfully induced cell differentiation in various AML cell lines. Through gene expression profiling analysis and differentiation marker genes CRISPR/Cas9 screening, the complement of TRIB1 and COP1 ubiquitin ligase complex was identified as a functional FOXO downstream gene that remains undifferentiated. TRIB1 is a direct target of FOXO3, and the FOXO in the TRIB1 promoter binds to cis regulatory elements, namely the FOXO responsive element (FRE-T) in the TRIB1 promoter, playing a crucial role in differentiation blockade. Discovering that cis regulatory elements targeting the FOXO family are a therapeutic strategy for inducing AML cell differentiation.
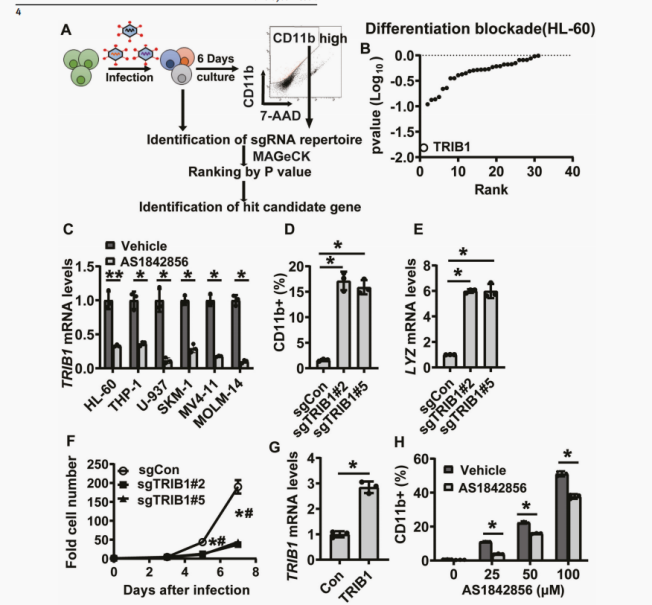
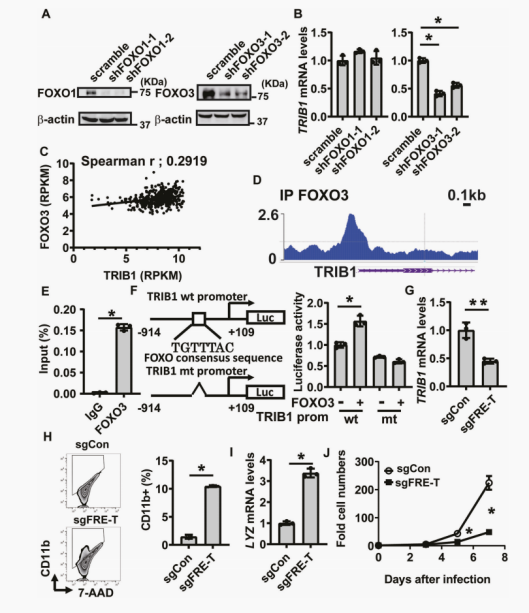
Down-regulation of TRIB1 through FOXO1 inhibitors contributes to the differentiation of AML cells, identifying cis regulatory elements of TRIB1 gene transcription to maintain the undifferentiated state of AML cells.
This study screened 30 genes to identify the key genes involved in AML differentiation blockade. And the validation of targets provides new ideas and methods for the treatment of leukemia.
EDITGENE provides a complete solution for CRISPR/Cas9 screening, including library design, plasmid library and lentivirus library generation, screening, NGS sequencing and MaGCKFlute analysis.
Reference:
1. SCARB2 drives hepatocellular carcinoma tumor initiating cells via enhanced MYC transcriptional activity
2. A membrane-associated MHC-I inhibitory axis for cancer immune evasion
3. Targeting cis-regulatory elements of FOXO family is a novel therapeutic strategy for induction of leukemia cell differentiation
info@editxor.com

